 |
PDBsum entry 2vcs

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2vcs
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.11.1.11
- L-ascorbate peroxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-ascorbate + H2O2 = L-dehydroascorbate + 2 H2O
|
 |
 |
 |
 |
 |
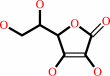 L-ascorbate
L-ascorbate
|
+
|
 H2O2
H2O2
|
=
|
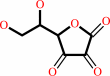 L-dehydroascorbate
L-dehydroascorbate
|
+
|
2
×
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme
|
 |
 |
 |
 |
 |
Heme
Bound ligand (Het Group name =
HEM)
matches with 95.45% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
283:6193
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
The tuberculosis prodrug isoniazid bound to activating peroxidases.
|
|
C.L.Metcalfe,
I.K.Macdonald,
E.J.Murphy,
K.A.Brown,
E.L.Raven,
P.C.Moody.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Isoniazid (INH, isonicotinic acid hydrazine) is one of only two therapeutic
agents effective in treating tuberculosis. This prodrug is activated by the heme
enzyme catalase-peroxidase (KatG) endogenous to Mycobacterium tuberculosis, but
the mechanism of activation is poorly understood, in part because the binding
interaction has not been properly established. The class I peroxidases ascorbate
peroxidase (APX) and cytochrome c peroxidase (CcP) have very similar active site
structures to KatG, and are also capable of activating isoniazid. We report here
the first crystal structures of complexes of isoniazid bound to APX and CcP.
These are the first structures of isoniazid bound to any activating enzymes. The
structures show that isoniazid binds close to the d-heme edge in both APX and
CcP, although the precise binding orientation varies slightly in the two cases.
A second binding site for INH is found in APX at the g-heme edge close to the
established ascorbate binding site, indicating that the g-heme edge can also
support the binding of aromatic substrates. We also show that in an active site
mutant of sAPX (W41A) INH can bind directly to the heme iron to become an
inhibitor, and in a different mode when the distal histidine is replaced by
alanine(H42A). These structures provide the first unambiguous evidence for the
location of the isoniazid binding site in the class I peroxidases, and provide
rationalisation of isoniazid resistance in naturally occurring KatG mutant
strains of M. tuberculosis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
FIGURE 4. Stereo diagrams showing INH bound in the active
site mutants of sAPX W41A and H42A. A, in sAPX·(W41A),
two molecules of INH (brown) are bound in the distal cavity, one
in the same position as sAPX and a second coordinated directly
to the heme iron. The figure shows observed F[o] - F[c]
difference density (in green, contoured at 3  ). B, in sAPX (H42A)
the orientation of INH (brown) is rotated relative to the wild
type and is held in position by a hydrogen bond to Trp-41.
Observed F[o] - F[c] difference density is shown in green. In
both cases the occupancy of the INH is partial and shared with
water molecules that are represented as red spheres. ). B, in sAPX (H42A)
the orientation of INH (brown) is rotated relative to the wild
type and is held in position by a hydrogen bond to Trp-41.
Observed F[o] - F[c] difference density is shown in green. In
both cases the occupancy of the INH is partial and shared with
water molecules that are represented as red spheres.
|
 |
Figure 6.
FIGURE 6. Stereo diagram showing INH bound in the ascorbate
binding pocket of sAPX. Hydrogen bonding interactions are
observed between INH and Arg-172 and Lys-31 and via a water to
the propionate group of the heme. There are INH molecules bound
in identical positions in the sAPX (W41A) and sAPX (H42A)
structures. The first INH molecule bound in the distal cavity is
also shown to aid orientation. The protein is shown in green,
the heme group in blue, and INH in pink. Waters are represented
as red spheres. The observed F[o] - F[c] difference density
(contoured at 3  ) for the INH molecules
is shown in green. Figs. 2, 3, 4, 5, 6 were prepared with PyMOL
(22). ) for the INH molecules
is shown in green. Figs. 2, 3, 4, 5, 6 were prepared with PyMOL
(22).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2007,
283,
6193)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
P.Ascenzi,
A.Bolli,
A.di Masi,
G.R.Tundo,
G.Fanali,
M.Coletta,
and
M.Fasano
(2011).
Isoniazid and rifampicin inhibit allosterically heme binding to albumin and peroxynitrite isomerization by heme-albumin.
|
| |
J Biol Inorg Chem,
16,
97.
|
 |
|
|
|
|
 |
A.K.Singh,
R.P.Kumar,
N.Pandey,
N.Singh,
M.Sinha,
A.Bhushan,
P.Kaur,
S.Sharma,
and
T.P.Singh
(2010).
Mode of binding of the tuberculosis prodrug isoniazid to heme peroxidases: binding studies and crystal structure of bovine lactoperoxidase with isoniazid at 2.7 A resolution.
|
| |
J Biol Chem,
285,
1569-1576.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.E.Cade,
A.C.Dlouhy,
K.F.Medzihradszky,
S.P.Salas-Castillo,
and
R.A.Ghiladi
(2010).
Isoniazid-resistance conferring mutations in Mycobacterium tuberculosis KatG: catalase, peroxidase, and INH-NADH adduct formation activities.
|
| |
Protein Sci,
19,
458-474.
|
 |
|
|
|
|
 |
A.K.Singh,
N.Singh,
S.Sharma,
K.Shin,
M.Takase,
P.Kaur,
A.Srinivasan,
and
T.P.Singh
(2009).
Inhibition of lactoperoxidase by its own catalytic product: crystal structure of the hypothiocyanate-inhibited bovine lactoperoxidase at 2.3-A resolution.
|
| |
Biophys J,
96,
646-654.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Suarez,
K.Ranguelova,
J.P.Schelvis,
and
R.S.Magliozzo
(2009).
Antibiotic resistance in Mycobacterium tuberculosis: peroxidase intermediate bypass causes poor isoniazid activation by the S315G mutant of M. tuberculosis catalase-peroxidase (KatG).
|
| |
J Biol Chem,
284,
16146-16155.
|
 |
|
|
|
|
 |
X.Zhao,
S.Yu,
K.Ranguelova,
J.Suarez,
L.Metlitsky,
J.P.Schelvis,
and
R.S.Magliozzo
(2009).
Role of the oxyferrous heme intermediate and distal side adduct radical in the catalase activity of Mycobacterium tuberculosis KatG revealed by the W107F mutant.
|
| |
J Biol Chem,
284,
7030-7037.
|
 |
|
|
|
|
 |
K.Ranguelova,
J.Suarez,
R.S.Magliozzo,
and
R.P.Mason
(2008).
Spin trapping investigation of peroxide- and isoniazid-induced radicals in Mycobacterium tuberculosis catalase-peroxidase.
|
| |
Biochemistry,
47,
11377-11385.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

