
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|
 223 a.a.
223 a.a.
|
 |

|

|

|

|

|

|

|
 241 a.a.
241 a.a.
|
 |

|

|

|

|

|

|

|

 255 a.a.
255 a.a.
|
 |

|

|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Gene regulation
|
 |
|
Title:
|
 |
Ure2p in complex with glutathione
|
|
Structure:
|
 |
Ure2 protein. Chain: a, b, c, d. Engineered: yes
|
|
Source:
|
 |
Saccharomyces cerevisiae. Baker's yeast. Organism_taxid: 4932. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
2.50Å
|
R-factor:
|
0.207
|
R-free:
|
0.264
|
|
|
Authors:
|
 |
L.Bousset,H.Belrhali,R.Melki,S.Morera
|
Key ref:

|
 |
L.Bousset
et al.
(2001).
Crystal structures of the yeast prion Ure2p functional region in complex with glutathione and related compounds.
Biochemistry,
40,
13564-13573.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
19-Sep-01
|
Release date:
|
21-Dec-01
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P23202
(URE2_YEAST) -
Transcriptional regulator URE2 from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
354 a.a.
223 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains A, B, C, D:
E.C.1.11.1.9
- glutathione peroxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 glutathione + H2O2 = glutathione disulfide + 2 H2O
|
 |
 |
 |
 |
 |
2
×
glutathione
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 H2O2
H2O2
|
=
|
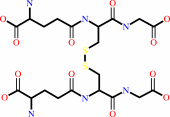 glutathione disulfide
glutathione disulfide
|
+
|
2
×
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Se(2+)
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B, C, D:
E.C.1.8.4.-
- ?????
|
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
40:13564-13573
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of the yeast prion Ure2p functional region in complex with glutathione and related compounds.
|
|
L.Bousset,
H.Belrhali,
R.Melki,
S.Morera.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The [URE3] phenotype in yeast Saccharomyces cerevisiae is due to an altered
prion form of Ure2p, a protein involved in nitrogen catabolism. To understand
possible conformational changes at the origin of prion propagation, we
previously solved the crystal structure of the Ure2p functional region [Bousset
et al. (2001) Structure 9, 39-46]. We showed the protein to have a fold similar
to that of the beta class of glutathione S-transferases (GSTs). Here we report
crystal structures of the Ure2p functional region (extending from residues
95-354) in complex with glutathione (GSH), the substrate of all GSTs, and two
widely used GST inhibitors, namely, S-hexylglutathione and
S-p-nitrobenzylglutathione. In a manner similar to what is observed in many
GSTs, ligand binding is not accompanied by a significant change in the
conformation of the protein. We identify one GSH and one hydrophobic
electrophile binding site per monomer as observed in all other GSTs. The sulfur
group of GSH, that conjugates electrophiles, is located near the amide group of
Asn124, allowing a hydrogen bond to be formed. Biochemical data indicate that
GSH binds to Ure2p with high affinity. Its binding affects Ure2p oligomerization
but has no effect on the assembly of the protein into amyloid fibrils. Despite
results indicating that Ure2p lacks GST activity, we propose that Ure2p is a
member of the GST superfamily that may describe a novel GST class. Our data
bring new insights into the function of the Ure2p active region.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.Chen,
L.J.Chen,
H.Y.Wang,
Y.Q.Wang,
and
S.Perrett
(2011).
Deletion of a Ure2 C-terminal prion-inhibiting region promotes the rate of fibril seed formation and alters interaction with Hsp40.
|
| |
Protein Eng Des Sel,
24,
69-78.
|
 |
|
|
|
|
 |
C.Zhang,
A.P.Jackson,
Z.R.Zhang,
Y.Han,
S.Yu,
R.Q.He,
and
S.Perrett
(2010).
Amyloid-like aggregates of the yeast prion protein ure2 enter vertebrate cells by specific endocytotic pathways and induce apoptosis.
|
| |
PLoS One,
5,
0.
|
 |
|
|
|
|
 |
L.Pieri,
M.Bucciantini,
P.Guasti,
J.Savistchenko,
R.Melki,
and
M.Stefani
(2009).
Synthetic lipid vesicles recruit native-like aggregates and affect the aggregation process of the prion Ure2p: insights on vesicle permeabilization and charge selectivity.
|
| |
Biophys J,
96,
3319-3330.
|
 |
|
|
|
|
 |
Z.R.Zhang,
and
S.Perrett
(2009).
Novel Glutaredoxin Activity of the Yeast Prion Protein Ure2 Reveals a Native-like Dimer within Fibrils.
|
| |
J Biol Chem,
284,
14058-14067.
|
 |
|
|
|
|
 |
B.Remmerie,
K.Vandenbroucke,
L.De Smet,
W.Carpentier,
D.De Vos,
J.Stout,
J.Van Beeumen,
and
S.N.Savvides
(2008).
Expression, purification, crystallization and structure determination of two glutathione S-transferase-like proteins from Shewanella oneidensis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
548-553.
|
 |
|
|
|
|
 |
J.Savistchenko,
J.Krzewska,
N.Fay,
and
R.Melki
(2008).
Molecular chaperones and the assembly of the prion Ure2p in vitro.
|
| |
J Biol Chem,
283,
15732-15739.
|
 |
|
|
|
|
 |
K.H.Wong,
M.J.Hynes,
and
M.A.Davis
(2008).
Recent advances in nitrogen regulation: a comparison between Saccharomyces cerevisiae and filamentous fungi.
|
| |
Eukaryot Cell,
7,
917-925.
|
 |
|
|
|
|
 |
F.Immel,
Y.Jiang,
Y.Q.Wang,
C.Marchal,
L.Maillet,
S.Perrett,
and
C.Cullin
(2007).
In vitro analysis of SpUre2p, a prion-related protein, exemplifies the relationship between amyloid and prion.
|
| |
J Biol Chem,
282,
7912-7920.
|
 |
|
|
|
|
 |
F.Shewmaker,
L.Mull,
T.Nakayashiki,
D.C.Masison,
and
R.B.Wickner
(2007).
Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae.
|
| |
Genetics,
176,
1557-1565.
|
 |
|
|
|
|
 |
H.Y.Lian,
H.Zhang,
Z.R.Zhang,
H.M.Loovers,
G.W.Jones,
P.J.Rowling,
L.S.Itzhaki,
J.M.Zhou,
and
S.Perrett
(2007).
Hsp40 interacts directly with the native state of the yeast prion protein Ure2 and inhibits formation of amyloid-like fibrils.
|
| |
J Biol Chem,
282,
11931-11940.
|
 |
|
|
|
|
 |
L.Pieri,
M.Bucciantini,
D.Nosi,
L.Formigli,
J.Savistchenko,
R.Melki,
and
M.Stefani
(2006).
The yeast prion Ure2p native-like assemblies are toxic to mammalian cells regardless of their aggregation state.
|
| |
J Biol Chem,
281,
15337-15344.
|
 |
|
|
|
|
 |
N.Ranson,
T.Stromer,
L.Bousset,
R.Melki,
and
L.C.Serpell
(2006).
Insights into the architecture of the Ure2p yeast protein assemblies from helical twisted fibrils.
|
| |
Protein Sci,
15,
2481-2487.
|
 |
|
|
|
|
 |
R.Singh,
M.A.White,
K.V.Ramana,
J.M.Petrash,
S.J.Watowich,
A.Bhatnagar,
and
S.K.Srivastava
(2006).
Structure of a glutathione conjugate bound to the active site of aldose reductase.
|
| |
Proteins,
64,
101-110.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.M.Hansen,
Y.Gu,
M.Li,
M.Andrykovitch,
D.S.Waugh,
D.J.Jin,
and
X.Ji
(2005).
Structural basis for the function of stringent starvation protein a as a transcription factor.
|
| |
J Biol Chem,
280,
17380-17391.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.D.Ross,
A.Minton,
and
R.B.Wickner
(2005).
Prion domains: sequences, structures and interactions.
|
| |
Nat Cell Biol,
7,
1039-1044.
|
 |
|
|
|
|
 |
N.Fay,
V.Redeker,
J.Savistchenko,
S.Dubois,
L.Bousset,
and
R.Melki
(2005).
Structure of the prion Ure2p in protein fibrils assembled in vitro.
|
| |
J Biol Chem,
280,
37149-37158.
|
 |
|
|
|
|
 |
S.Catharino,
J.Buchner,
and
S.Walter
(2005).
Characterization of oligomeric species in the fibrillization pathway of the yeast prion Ure2p.
|
| |
Biol Chem,
386,
633-641.
|
 |
|
|
|
|
 |
M.Bai,
J.M.Zhou,
and
S.Perrett
(2004).
The yeast prion protein Ure2 shows glutathione peroxidase activity in both native and fibrillar forms.
|
| |
J Biol Chem,
279,
50025-50030.
|
 |
|
|
|
|
 |
M.F.Tuite,
and
B.S.Cox
(2003).
Propagation of yeast prions.
|
| |
Nat Rev Mol Cell Biol,
4,
878-890.
|
 |
|
|
|
|
 |
R.Rai,
J.J.Tate,
and
T.G.Cooper
(2003).
Ure2, a prion precursor with homology to glutathione S-transferase, protects Saccharomyces cerevisiae cells from heavy metal ion and oxidant toxicity.
|
| |
J Biol Chem,
278,
12826-12833.
|
 |
|
|
|
|
 |
U.Baxa,
K.L.Taylor,
J.S.Wall,
M.N.Simon,
N.Cheng,
R.B.Wickner,
and
A.C.Steven
(2003).
Architecture of Ure2p prion filaments: the N-terminal domains form a central core fiber.
|
| |
J Biol Chem,
278,
43717-43727.
|
 |
|
|
|
|
 |
J.A.Fraser,
M.A.Davis,
and
M.J.Hynes
(2002).
A gene from Aspergillus nidulans with similarity to URE2 of Saccharomyces cerevisiae encodes a glutathione S-transferase which contributes to heavy metal and xenobiotic resistance.
|
| |
Appl Environ Microbiol,
68,
2802-2808.
|
 |
|
|
|
|
 |
L.Bousset,
N.H.Thomson,
S.E.Radford,
and
R.Melki
(2002).
The yeast prion Ure2p retains its native alpha-helical conformation upon assembly into protein fibrils in vitro.
|
| |
EMBO J,
21,
2903-2911.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |
|

