 |
PDBsum entry 1i1d

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.4
- glucosamine-phosphate N-acetyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
UDP-N-acetylglucosamine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-glucosamine 6-phosphate + acetyl-CoA = N-acetyl-D-glucosamine 6-phosphate + CoA + H+
|
 |
 |
 |
 |
 |
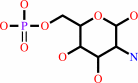 D-glucosamine 6-phosphate
D-glucosamine 6-phosphate
|
+
|
 acetyl-CoA
acetyl-CoA
|
=
|
N-acetyl-D-glucosamine 6-phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 CoA
CoA
|
+
|
H(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
276:16328-16334
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
The crystal structures of Apo and complexed Saccharomyces cerevisiae GNA1 shed light on the catalytic mechanism of an amino-sugar N-acetyltransferase.
|
|
C.Peneff,
D.Mengin-Lecreulx,
Y.Bourne.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The yeast enzymes involved in UDP-GlcNAc biosynthesis are potential targets for
antifungal agents. GNA1, a novel member of the Gcn5-related N-acetyltransferase
(GNAT) superfamily, participates in UDP-GlcNAc biosynthesis by catalyzing the
formation of GlcNAc6P from AcCoA and GlcN6P. We have solved three crystal
structures corresponding to the apo Saccharomyces cerevisiae GNA1, the
GNA1-AcCoA, and the GNA1-CoA-GlcNAc6P complexes and have refined them to 2.4,
1.3, and 1.8 A resolution, respectively. These structures not only reveal a
stable, beta-intertwined, dimeric assembly with the GlcNAc6P binding site
located at the dimer interface but also shed light on the catalytic machinery of
GNA1 at an atomic level. Hence, they broaden our understanding of structural
features required for GNAT activity, provide structural details for related
aminoglycoside N-acetyltransferases, and highlight the adaptability of the GNAT
superfamily members to acquire various specificities.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Schematic representation of the proposed GNA1
catalytic mechanism.
|
 |
Figure 5.
Fig. 5. Structural comparison of substrate binding sites
of AANAT , tGCN5, and GNA1. The molecular surfaces around the
active site of the AANAT-bisubstrate analog (A), the
tGCN5-CoA-H3peptide (B), and the GNA1-CoA-GlcNAc6P complex
structures (C) are viewed with a similar orientation. From top
to bottom, the serotonin-like moiety and the acetyl group are
shown in red, the H3 peptide backbone in yellow, with its
reactive Lys-14 side chain in orange, and GlcNAc6P in purple.
Structural divergences within the NH[2]- and COOH-terminal
regions are highlighted in dark blue and blue, respectively. The
 3- 3-  4 insertion
loop unique to AANAT is shown in brown. 4 insertion
loop unique to AANAT is shown in brown.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2001,
276,
16328-16334)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.C.Dorfmueller,
W.Fang,
F.V.Rao,
D.E.Blair,
H.Attrill,
and
D.M.van Aalten
(2012).
Structural and biochemical characterization of a trapped coenzyme A adduct of Caenorhabditis elegans glucosamine-6-phosphate N-acetyltransferase 1.
|
| |
Acta Crystallogr D Biol Crystallogr,
68,
1019-1029.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Shameer,
G.Pugalenthi,
K.K.Kandaswamy,
P.N.Suganthan,
G.Archunan,
and
R.Sowdhamini
(2010).
Insights into Protein Sequence and Structure-Derived Features Mediating 3D Domain Swapping Mechanism using Support Vector Machine Based Approach.
|
| |
Bioinform Biol Insights,
4,
33-42.
|
 |
|
|
|
|
 |
A.M.Davies,
R.Tata,
F.X.Chauviac,
B.J.Sutton,
and
P.R.Brown
(2008).
Structure of a putative acetyltransferase (PA1377) from Pseudomonas aeruginosa.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
338-342.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Barreteau,
A.Kovac,
A.Boniface,
M.Sova,
S.Gobec,
and
D.Blanot
(2008).
Cytoplasmic steps of peptidoglycan biosynthesis.
|
| |
FEMS Microbiol Rev,
32,
168-207.
|
 |
|
|
|
|
 |
T.Kotani,
and
H.Takagi
(2008).
Identification of amino acid residues essential for the yeast N-acetyltransferase Mpr1 activity by site-directed mutagenesis.
|
| |
FEMS Yeast Res,
8,
607-614.
|
 |
|
|
|
|
 |
W.Zhang,
V.C.Jones,
M.S.Scherman,
S.Mahapatra,
D.Crick,
S.Bhamidi,
Y.Xin,
M.R.McNeil,
and
Y.Ma
(2008).
Expression, essentiality, and a microtiter plate assay for mycobacterial GlmU, the bifunctional glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase.
|
| |
Int J Biochem Cell Biol,
40,
2560-2571.
|
 |
|
|
|
|
 |
J.Wang,
Y.F.Zhou,
L.F.Li,
Y.H.Liang,
and
X.D.Su
(2006).
Purification, crystallization and preliminary X-ray analysis of the glucosamine-6-phosphate N-acetyltransferase from human liver.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
1097-1099.
|
 |
|
|
|
|
 |
M.N.Hung,
E.Rangarajan,
C.Munger,
G.Nadeau,
T.Sulea,
and
A.Matte
(2006).
Crystal structure of TDP-fucosamine acetyltransferase (WecD) from Escherichia coli, an enzyme required for enterobacterial common antigen synthesis.
|
| |
J Bacteriol,
188,
5606-5617.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.M.Van Wagoner,
and
J.Clardy
(2006).
FeeM, an N-acyl amino acid synthase from an uncultured soil microbe: structure, mechanism, and acyl carrier protein binding.
|
| |
Structure,
14,
1425-1435.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Milewski,
I.Gabriel,
and
J.Olchowy
(2006).
Enzymes of UDP-GlcNAc biosynthesis in yeast.
|
| |
Yeast,
23,
1.
|
 |
|
|
|
|
 |
Y.Hahn,
and
B.Lee
(2006).
Human-specific nonsense mutations identified by genome sequence comparisons.
|
| |
Hum Genet,
119,
169-178.
|
 |
|
|
|
|
 |
D.L.Burk,
B.Xiong,
C.Breitbach,
and
A.M.Berghuis
(2005).
Structures of aminoglycoside acetyltransferase AAC(6')-Ii in a novel crystal form: structural and normal-mode analyses.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1273-1279.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.S.Brunzelle,
R.Wu,
S.V.Korolev,
F.R.Collart,
A.Joachimiak,
and
W.F.Anderson
(2004).
Crystal structure of Bacillus subtilis YdaF protein: a putative ribosomal N-acetyltransferase.
|
| |
Proteins,
57,
850-853.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Behnia,
B.Panic,
J.R.Whyte,
and
S.Munro
(2004).
Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p.
|
| |
Nat Cell Biol,
6,
405-413.
|
 |
|
|
|
|
 |
R.Palanimurugan,
H.Scheel,
K.Hofmann,
and
R.J.Dohmen
(2004).
Polyamines regulate their synthesis by inducing expression and blocking degradation of ODC antizyme.
|
| |
EMBO J,
23,
4857-4867.
|
 |
|
|
|
|
 |
S.Sarkhel,
and
G.R.Desiraju
(2004).
N-H...O, O-H...O, and C-H...O hydrogen bonds in protein-ligand complexes: strong and weak interactions in molecular recognition.
|
| |
Proteins,
54,
247-259.
|
 |
|
|
|
|
 |
B.Taneja,
S.Maar,
L.Shuvalova,
F.Collart,
W.Anderson,
and
A.Mondragón
(2003).
Structure of the bacillus subtilis YYCN protein: a putative N-acetyltransferase.
|
| |
Proteins,
53,
950-952.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.W.Vetting,
S.L.Roderick,
M.Yu,
and
J.S.Blanchard
(2003).
Crystal structure of mycothiol synthase (Rv0819) from Mycobacterium tuberculosis shows structural homology to the GNAT family of N-acetyltransferases.
|
| |
Protein Sci,
12,
1954-1959.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.W.Vetting,
S.S.Hegde,
F.Javid-Majd,
J.S.Blanchard,
and
S.L.Roderick
(2002).
Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates.
|
| |
Nat Struct Biol,
9,
653-658.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

