 |
PDBsum entry 1gqg

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1gqg
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Quercetin 2,3-dioxygenase in complex with the inhibitor diethyldithiocarbamate
|
|
Structure:
|
 |
Quercetin 2,3-dioxygenase. Chain: a, b, c, d. Engineered: yes
|
|
Source:
|
 |
Aspergillus japonicus. Organism_taxid: 34381. Expressed in: aspergillus awamori. Expression_system_taxid: 105351
|
|
Biol. unit:
|
 |
 Dimer (from PDB file)
Dimer (from PDB file)
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.162
|
R-free:
|
0.183
|
|
|
Authors:
|
 |
R.A.Steiner,B.W.Dijkstra
|
Key ref:

|
 |
R.A.Steiner
et al.
(2002).
Functional analysis of the copper-dependent quercetin 2,3-dioxygenase. 1. Ligand-induced coordination changes probed by X-ray crystallography: inhibition, ordering effect, and mechanistic insights.
Biochemistry,
41,
7955-7962.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
23-Nov-01
|
Release date:
|
21-Jun-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q7SIC2
(QDOI_ASPJA) -
Quercetin 2,3-dioxygenase from Aspergillus japonicus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
350 a.a.
333 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.13.11.24
- quercetin 2,3-dioxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Quercetin 2,3-Dioxygenase
|
 |
 |
 |
 |
 |
Reaction:
|
 |
quercetin + O2 = 2-(3,4-dihydroxybenzoyloxy)-4,6-dihydroxybenzoate + CO
|
 |
 |
 |
 |
 |
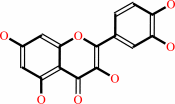 quercetin
quercetin
|
+
|
O2
Bound ligand (Het Group name = )
matches with 43.48% similarity
|
=
|
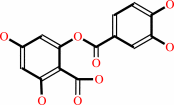 2-(3,4-dihydroxybenzoyloxy)-4,6-dihydroxybenzoate
2-(3,4-dihydroxybenzoyloxy)-4,6-dihydroxybenzoate
|
+
|
 CO
CO
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe cation or Cu cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
41:7955-7962
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Functional analysis of the copper-dependent quercetin 2,3-dioxygenase. 1. Ligand-induced coordination changes probed by X-ray crystallography: inhibition, ordering effect, and mechanistic insights.
|
|
R.A.Steiner,
I.M.Kooter,
B.W.Dijkstra.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structures of the copper-dependent Aspergillus japonicus quercetin
2,3-dioxygenase (2,3QD) complexed with the inhibitors diethyldithiocarbamate
(DDC) and kojic acid (KOJ) are reported at 1.70 and 2.15 A resolution,
respectively. Both inhibitors asymmetrically chelate the metal center and assume
a common orientation in the active site cleft. Their molecular plane blocks
access to the inner portion of the cavity which is lined by the side chains of
residues Met51, Thr53, Phe75, Phe114, and Met123 and which is believed to bind
the flavonol B-ring of the natural substrate. The binding of the inhibitors
brings order into the mixed coordination observed in the native enzyme. DDC and
KOJ induce a single conformation of the Glu73 side chain, although in different
ways. In the presence of DDC, Glu73 is detached from the copper ion with its
carboxylate moiety pointing away from the active site cavity. In contrast, when
KOJ is bound, Glu73 ligates the Cu ion through its O(epsilon)(1) atom with a
monodentate geometry. Compared to the native coordinating conformation, this
conformation is approximately 90 degrees rotated about the chi(3) angle. This
latter Glu73 conformation is compatible with the presence of a bound substrate.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Miyazaki,
S.Simizu,
H.Okumura,
S.Takagi,
and
H.Osada
(2010).
A small-molecule inhibitor shows that pirin regulates migration of melanoma cells.
|
| |
Nat Chem Biol,
6,
667-673.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Tranchimand,
P.Brouant,
and
G.Iacazio
(2010).
The rutin catabolic pathway with special emphasis on quercetinase.
|
| |
Biodegradation,
21,
833-859.
|
 |
|
|
|
|
 |
I.Matera,
M.Ferraroni,
S.Bürger,
A.Stolz,
and
F.Briganti
(2006).
Preliminary crystallographic analysis of salicylate 1,2-dioxygenase from Pseudaminobacter salicylatoxidans.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
553-555.
|
 |
|
|
|
|
 |
R.Bentley
(2006).
From miso, saké and shoyu to cosmetics: a century of science for kojic acid.
|
| |
Nat Prod Rep,
23,
1046-1062.
|
 |
|
|
|
|
 |
D.G.Covell,
A.Wallqvist,
R.Huang,
N.Thanki,
A.A.Rabow,
and
X.J.Lu
(2005).
Linking tumor cell cytotoxicity to mechanism of drug action: an integrated analysis of gene expression, small-molecule screening and structural databases.
|
| |
Proteins,
59,
403-433.
|
 |
|
|
|
|
 |
M.Adams,
and
Z.Jia
(2005).
Structural and biochemical analysis reveal pirins to possess quercetinase activity.
|
| |
J Biol Chem,
280,
28675-28682.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Raymond,
A.Tocilj,
E.Ajamian,
Y.Li,
M.N.Hung,
A.Matte,
and
M.Cygler
(2005).
Crystal structure of ureidoglycolate hydrolase (AllA) from Escherichia coli O157:H7.
|
| |
Proteins,
61,
454-459.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Fittipaldi,
R.A.Steiner,
M.Matsushita,
B.W.Dijkstra,
E.J.Groenen,
and
M.Huber
(2003).
Single-crystal EPR study at 95 GHz of the type 2 copper site of the inhibitor-bound quercetin 2,3-dioxygenase.
|
| |
Biophys J,
85,
4047-4054.
|
 |
|
|
|
|
 |
I.M.Kooter,
R.A.Steiner,
B.W.Dijkstra,
P.I.van Noort,
M.R.Egmond,
and
M.Huber
(2002).
EPR characterization of the mononuclear Cu-containing Aspergillus japonicus quercetin 2,3-dioxygenase reveals dramatic changes upon anaerobic binding of substrates.
|
| |
Eur J Biochem,
269,
2971-2979.
|
 |
|
|
|
|
 |
R.A.Steiner,
K.H.Kalk,
and
B.W.Dijkstra
(2002).
Anaerobic enzyme.substrate structures provide insight into the reaction mechanism of the copper-dependent quercetin 2,3-dioxygenase.
|
| |
Proc Natl Acad Sci U S A,
99,
16625-16630.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

