 |
PDBsum entry 1e2j

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.21
- thymidine kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
thymidine + ATP = dTMP + ADP + H+
|
 |
 |
 |
 |
 |
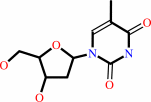 thymidine
thymidine
|
+
|
ATP
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 dTMP
dTMP
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
41:545-553
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Nucleoside binding site of herpes simplex type 1 thymidine kinase analyzed by X-ray crystallography.
|
|
J.Vogt,
R.Perozzo,
A.Pautsch,
A.Prota,
P.Schelling,
B.Pilger,
G.Folkers,
L.Scapozza,
G.E.Schulz.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structures of the full-length Herpes simplex virus type 1 thymidine
kinase in its unligated form and in a complex with an adenine analogue have been
determined at 1.9 A resolution. The unligated enzyme contains four water
molecules in the thymidine pocket and reveals a small induced fit on substrate
binding. The structure of the ligated enzyme shows for the first time a bound
adenine analogue after numerous complexes with thymine and guanine analogues
have been reported. The adenine analogue constitutes a new lead compound for
enzyme-prodrug gene therapy. In addition, the structure of mutant Q125N
modifying the binding site of the natural substrate thymidine in complex with
this substrate has been established at 2.5 A resolution. It reveals that neither
the binding mode of thymidine nor the polypeptide backbone conformation is
altered, except that the two major hydrogen bonds to thymidine are replaced by a
single water-mediated hydrogen bond, which improves the relative acceptance of
the prodrugs aciclovir and ganciclovir compared with the natural substrate.
Accordingly, the mutant structure represents a first step toward improving the
virus-directed enzyme-prodrug gene therapy by enzyme engineering.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. Structural formulae of the adenosine analogue
9-(2-(S)-hydroxypropyl)adenine (HPA) and the AMP analogue
9-(2-phosphonylmethoxyethyl)adenine (PMEA). HPA was used as a
racemic mixture.
|
 |
Figure 6.
Figure 6. Superposition of mutant TK[HSV1](Q125N) in complex
with dT (thick solid lines, C  atoms
marked by small dots, hydrogen bonds are dotted) with wild-type
TK[HSV1] in complex with aciclovir (thin solid lines),[19]
ganciclovir (thin dotted lines)[19]and dT (thick dotted
lines).[19] For sake of clarity, side chains are only drawn for
TK[HSV1](Q125N):dT and TK[HSV1]:aciclovir. Water molecules and
hydrogen bonds are depicted for TK[HSV1](Q125N):dT (black
circles) and TK[HSV1]:aciclovir (pale circles). For aciclovir,
only the location of the hydroxyl group at the 5 atoms
marked by small dots, hydrogen bonds are dotted) with wild-type
TK[HSV1] in complex with aciclovir (thin solid lines),[19]
ganciclovir (thin dotted lines)[19]and dT (thick dotted
lines).[19] For sake of clarity, side chains are only drawn for
TK[HSV1](Q125N):dT and TK[HSV1]:aciclovir. Water molecules and
hydrogen bonds are depicted for TK[HSV1](Q125N):dT (black
circles) and TK[HSV1]:aciclovir (pale circles). For aciclovir,
only the location of the hydroxyl group at the 5  -OH
position of dT is shown. -OH
position of dT is shown.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2000,
41,
545-553)
copyright 2000.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.I.Sułkowska,
P.Sułkowski,
and
J.N.Onuchic
(2009).
Jamming proteins with slipknots and their free energy landscape.
|
| |
Phys Rev Lett,
103,
268103.
|
 |
|
|
|
|
 |
L.Egeblad-Welin,
Y.Sonntag,
H.Eklund,
and
B.Munch-Petersen
(2007).
Functional studies of active-site mutants from Drosophila melanogaster deoxyribonucleoside kinase. Investigations of the putative catalytic glutamate-arginine pair and of residues responsible for substrate specificity.
|
| |
FEBS J,
274,
1542-1551.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.T.Gammon,
M.Bernstein,
D.P.Schuster,
and
D.Piwnica-Worms
(2007).
A method for quantification of nucleotides and nucleotide analogues in thymidine kinase assays using lanthanum phosphate coprecipitation.
|
| |
Anal Biochem,
369,
80-86.
|
 |
|
|
|
|
 |
C.A.Bottoms,
T.A.White,
and
J.J.Tanner
(2006).
Exploring structurally conserved solvent sites in protein families.
|
| |
Proteins,
64,
404-421.
|
 |
|
|
|
|
 |
J.Balzarini,
S.Liekens,
N.Solaroli,
K.El Omari,
D.K.Stammers,
and
A.Karlsson
(2006).
Engineering of a single conserved amino acid residue of herpes simplex virus type 1 thymidine kinase allows a predominant shift from pyrimidine to purine nucleoside phosphorylation.
|
| |
J Biol Chem,
281,
19273-19279.
|
 |
|
|
|
|
 |
K.El Omari,
N.Solaroli,
A.Karlsson,
J.Balzarini,
and
D.K.Stammers
(2006).
Structure of vaccinia virus thymidine kinase in complex with dTTP: insights for drug design.
|
| |
BMC Struct Biol,
6,
22.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Kotaka,
B.Dhaliwal,
J.Ren,
C.E.Nichols,
R.Angell,
M.Lockyer,
A.R.Hawkins,
and
D.K.Stammers
(2006).
Structures of S. aureus thymidylate kinase reveal an atypical active site configuration and an intermediate conformational state upon substrate binding.
|
| |
Protein Sci,
15,
774-784.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Haouz,
V.Vanheusden,
H.Munier-Lehmann,
M.Froeyen,
P.Herdewijn,
S.Van Calenbergh,
and
M.Delarue
(2003).
Enzymatic and structural analysis of inhibitors designed against Mycobacterium tuberculosis thymidylate kinase. New insights into the phosphoryl transfer mechanism.
|
| |
J Biol Chem,
278,
4963-4971.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.E.Bird,
J.Ren,
A.Wright,
K.D.Leslie,
B.Degrève,
J.Balzarini,
and
D.K.Stammers
(2003).
Crystal structure of varicella zoster virus thymidine kinase.
|
| |
J Biol Chem,
278,
24680-24687.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Wurth,
U.Kessler,
J.Vogt,
G.E.Schulz,
G.Folkers,
and
L.Scapozza
(2001).
The effect of substrate binding on the conformation and structural stability of Herpes simplex virus type 1 thymidine kinase.
|
| |
Protein Sci,
10,
63-73.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

