 |
PDBsum entry 7zzv

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Protein fibril
|
PDB id
|
|

|

|
7zzv
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Protein fibril
|
 |
|
Title:
|
 |
Prostatic acid phosphatase (pap) fragment (85-120)
|
|
Structure:
|
 |
Prostatic acid phosphatase. Chain: a. Synonym: pap,5'-nucleotidase,5'-nt,acid phosphatase 3,ecto-5'- nucleotidase,protein tyrosine phosphatase acp3,thiamine monophosphatase,tmpase. Engineered: yes
|
|
Source:
|
 |
Synthetic: yes. Homo sapiens. Human. Organism_taxid: 9606
|
|
NMR struc:
|
 |
18 models
|
 |
|
Authors:
|
 |
D.S.Blokhin,A.R.Yulmetov,A.M.Kusova,P.V.Skvortsova
|
|
Key ref:
|
 |
D.S.Blokhin
et al.
(2022).
Structure of amyloidogenic pap(85-120) peptide by high-Resolution nmr spectroscopy.
Journal of molecular,
1253,
.
|
 |
|
Date:
|
 |
|
26-May-22
|
Release date:
|
15-Jun-22
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P15309
(PPAP_HUMAN) -
Prostatic acid phosphatase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
386 a.a.
36 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.1.3.2
- acid phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a phosphate monoester + H2O = an alcohol + phosphate
|
 |
 |
 |
 |
 |
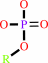 phosphate monoester
phosphate monoester
|
+
|
H2O
|
=
|
 alcohol
alcohol
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.1.3.48
- protein-tyrosine-phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
O-phospho-L-tyrosyl-[protein] + H2O = L-tyrosyl-[protein] + phosphate
|
 |
 |
 |
 |
 |
O-phospho-L-tyrosyl-[protein]
|
+
|
H2O
|
=
|
L-tyrosyl-[protein]
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.1.3.5
- 5'-nucleotidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a ribonucleoside 5'-phosphate + H2O = a ribonucleoside + phosphate
|
 |
 |
 |
 |
 |
ribonucleoside 5'-phosphate
|
+
|
H2O
|
=
|
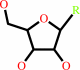 ribonucleoside
ribonucleoside
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

