 |
PDBsum entry 7f9c

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Ligase/ligase inhibitor
|
PDB id
|
|

|

|
7f9c
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase/ligase inhibitor
|
 |
|
Title:
|
 |
Homo sapiens prolyl-tRNA synthetase (hsprs) in complex with l-proline and compound l96
|
|
Structure:
|
 |
Bifunctional glutamate/proline--tRNA ligase. Chain: a, b. Synonym: bifunctional aminoacyl-tRNA synthetase,cell proliferation- inducing gene 32 protein,glutamatyl-prolyl-tRNA synthetase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: eprs1, eprs, glns, pars, qars, qprs, pig32. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.191
|
R-free:
|
0.253
|
|
|
Authors:
|
 |
N.Malhotra,Y.Manickam,A.Sharma
|
|
Key ref:
|
 |
N.Malhotra
et al.
Hsprs with ATP binding pocket inhibitor.
To be published,
.
PubMed id:
|
 |
|
Date:
|
 |
|
04-Jul-21
|
Release date:
|
06-Jul-22
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P07814
(SYEP_HUMAN) -
Bifunctional glutamate/proline--tRNA ligase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1512 a.a.
488 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.6.1.1.15
- proline--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Pro) + L-proline + ATP = L-prolyl-tRNA(Pro) + AMP + diphosphate
|
 |
 |
 |
 |
 |
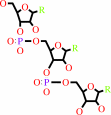 tRNA(Pro)
tRNA(Pro)
|
+
|
L-proline
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 ATP
ATP
|
=
|
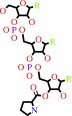 L-prolyl-tRNA(Pro)
L-prolyl-tRNA(Pro)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.6.1.1.17
- glutamate--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Glu) + L-glutamate + ATP = L-glutamyl-tRNA(Glu) + AMP + diphosphate
|
 |
 |
 |
 |
 |
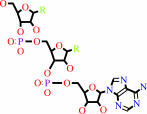 tRNA(Glu)
tRNA(Glu)
|
+
|
L-glutamate
Bound ligand (Het Group name = )
matches with 80.00% similarity
|
+
|
 ATP
ATP
|
=
|
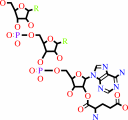 L-glutamyl-tRNA(Glu)
L-glutamyl-tRNA(Glu)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

