 |
PDBsum entry 5u3f
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.6.1.42
- branched-chain-amino-acid transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Leucine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-leucine + 2-oxoglutarate = 4-methyl-2-oxopentanoate + L-glutamate
|
 |
 |
 |
 |
 |
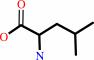 L-leucine
L-leucine
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
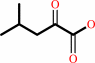 4-methyl-2-oxopentanoate
4-methyl-2-oxopentanoate
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
7TS)
matches with 65.22% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
ACS Chem Biol
12:1235-1244
(2017)
|
|
PubMed id:
|
|
|
|

|
| |
|
Mechanism-Based Inhibition of the Mycobacterium tuberculosis Branched-Chain Aminotransferase by d- and l-Cycloserine.
|
|
T.M.Amorim Franco,
L.Favrot,
O.Vergnolle,
J.S.Blanchard.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The branched-chain aminotransferase is a pyridoxal 5'-phosphate (PLP)-dependent
enzyme responsible for the final step in the biosynthesis of all three
branched-chain amino acids, l-leucine, l-isoleucine, and l-valine, in bacteria.
We have investigated the mechanism of inactivation of the branched-chain
aminotransferase from Mycobacterium tuberculosis (MtIlvE) by d- and
l-cycloserine. d-Cycloserine is currently used only in the treatment of
multidrug-drug-resistant tuberculosis. Our results show a time- and
concentration-dependent inactivation of MtIlvE by both isomers, with
l-cycloserine being a 40-fold better inhibitor of the enzyme. Minimum inhibitory
concentration (MIC) studies revealed that l-cycloserine is a 10-fold better
inhibitor of Mycobacterium tuberculosis growth than d-cycloserine. In addition,
we have crystallized the MtIlvE-d-cycloserine inhibited enzyme, determining the
structure to 1.7 Å. The structure of the covalent d-cycloserine-PMP adduct
bound to MtIlvE reveals that the d-cycloserine ring is planar and aromatic, as
previously observed for other enzyme systems. Mass spectrometry reveals that
both the d-cycloserine- and l-cycloserine-PMP complexes have the same mass, and
are likely to be the same aromatized, isoxazole product. However, the kinetics
of formation of the MtIlvE d-cycloserine-PMP and MtIlvE l-cycloserine-PMP
adducts are quite different. While the kinetics of the formation of the MtIlvE
d-cycloserine-PMP complex can be fit to a single exponential, the formation of
the MtIlvE l-cycloserine-PMP complex occurs in two steps. We propose a chemical
mechanism for the inactivation of d- and l-cycloserine which suggests a
stereochemically determined structural role for the differing kinetics of
inactivation. These results demonstrate that the mechanism of action of
d-cycloserine's activity against M. tuberculosis may be more complicated than
previously thought and that d-cycloserine may compromise the in vivo activity of
multiple PLP-dependent enzymes, including MtIlvE.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

