 |
PDBsum entry 5mcp

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
5mcp
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|



 440 a.a.
440 a.a.
|
 |

|

|

|

|

|

|

|
 387 a.a.
387 a.a.
|
 |

|

|

|

|

|

|

|
 342 a.a.
342 a.a.
|
 |

|

|

|

|

|

|

|
 382 a.a.
382 a.a.
|
 |

|

|

|

|

|

|

|
 321 a.a.
321 a.a.
|
 |

|

|
|
|
|
|
|
|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Structure of imp dehydrogenase from ashbya gossypii bound to atp
|
|
Structure:
|
 |
Inosine-5'-monophosphate dehydrogenase. Chain: a, b, c, d, e, f, g, h. Synonym: impdh. Engineered: yes
|
|
Source:
|
 |
Ashbya gossypii (strain atcc 10895 / cbs 109.51 / fgsc 9923 / nrrl y-1056). Yeast. Organism_taxid: 284811. Strain: atcc 10895 / cbs 109.51 / fgsc 9923 / nrrl y-1056. Gene: agos_aer117w. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.251
|
R-free:
|
0.271
|
|
|
Authors:
|
 |
G.Winter,D.Fernandez-Justel,J.M.De Pereda,J.L.Revuelta,R.M.Buey
|
|
Key ref:
|
 |
R.M.Buey
et al.
(2017).
A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases.
Sci Rep,
7,
2648.
PubMed id:
|
 |
|
Date:
|
 |
|
10-Nov-16
|
Release date:
|
14-Jun-17
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q756Z6
(Q756Z6_ASHGO) -
Inosine-5'-monophosphate dehydrogenase from Eremothecium gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
522 a.a.
440 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
Q756Z6
(Q756Z6_ASHGO) -
Inosine-5'-monophosphate dehydrogenase from Eremothecium gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
522 a.a.
387 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
Q756Z6
(Q756Z6_ASHGO) -
Inosine-5'-monophosphate dehydrogenase from Eremothecium gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
522 a.a.
342 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F, G, H:
E.C.1.1.1.205
- Imp dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP and GMP Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + NAD+ + H2O = XMP + NADH + H+
|
 |
 |
 |
 |
 |
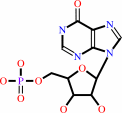 IMP
IMP
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 68.75% similarity
|
+
|
H2O
|
=
|
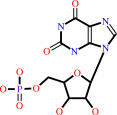 XMP
XMP
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Sci Rep
7:2648
(2017)
|
|
PubMed id:
|
|
|
|

|
| |
|
A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases.
|
|
R.M.Buey,
D.Fernández-Justel,
�.�.Marcos-Alcalde,
G.Winter,
P.Gómez-Puertas,
J.M.de Pereda,
J.Luis Revuelta.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Inosine-5'-monophosphate dehydrogenase (IMPDH) is an essential enzyme for
nucleotide metabolism and cell proliferation. Despite IMPDH is the target of
drugs with antiviral, immunosuppressive and antitumor activities, its
physiological mechanisms of regulation remain largely unknown. Using the enzyme
from the industrial fungus Ashbya gossypii, we demonstrate that the binding of
adenine and guanine nucleotides to the canonical nucleotide binding sites of the
regulatory Bateman domain induces different enzyme conformations with
significantly distinct catalytic activities. Thereby, the comparison of their
high-resolution structures defines the mechanistic and structural details of a
nucleotide-controlled conformational switch that allosterically modulates the
catalytic activity of eukaryotic IMPDHs. Remarkably, retinopathy-associated
mutations lie within the mechanical hinges of the conformational change,
highlighting its physiological relevance. Our results expand the mechanistic
repertoire of Bateman domains and pave the road to new approaches targeting
IMPDHs.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |

| | |

