 |
PDBsum entry 5k4x

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
5k4x
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.205
- Imp dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP and GMP Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + NAD+ + H2O = XMP + NADH + H+
|
 |
 |
 |
 |
 |
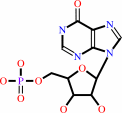 IMP
IMP
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H2O
|
=
|
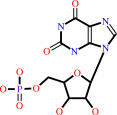 XMP
XMP
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acs Infect Dis
3:18-33
(2017)
|
|
PubMed id:
|
|
|
|

|
| |
|
Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
|
|
Y.Park,
A.Pacitto,
T.Bayliss,
L.A.Cleghorn,
Z.Wang,
T.Hartman,
K.Arora,
T.R.Ioerger,
J.Sacchettini,
M.Rizzi,
S.Donini,
T.L.Blundell,
D.B.Ascher,
K.Rhee,
A.Breda,
N.Zhou,
V.Dartois,
S.R.Jonnala,
L.E.Via,
V.Mizrahi,
O.Epemolu,
L.Stojanovski,
F.Simeons,
M.Osuna-Cabello,
L.Ellis,
C.J.MacKenzie,
A.R.Smith,
S.H.Davis,
D.Murugesan,
K.I.Buchanan,
P.A.Turner,
M.Huggett,
F.Zuccotto,
M.J.Rebollo-Lopez,
M.J.Lafuente-Monasterio,
O.Sanz,
G.S.Diaz,
J.Lelièvre,
L.Ballell,
C.Selenski,
M.Axtman,
S.Ghidelli-Disse,
H.Pflaumer,
M.Bösche,
G.Drewes,
G.M.Freiberg,
M.D.Kurnick,
M.Srikumaran,
D.J.Kempf,
S.R.Green,
P.C.Ray,
K.Read,
P.Wyatt,
C.E.Barry,
H.I.Boshoff.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

