 |
PDBsum entry 4zk7

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Protein binding
|
PDB id
|
|

|

|
4zk7
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|





 (+ 6 more)
117 a.a.
(+ 6 more)
117 a.a.
|
 |

|

|

|

|

|

|

|





 (+ 5 more)
103 a.a.
(+ 5 more)
103 a.a.
|
 |

|

|

|

|

|

|

|
 109 a.a.
109 a.a.
|
 |

|

|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Protein binding
|
 |
|
Title:
|
 |
Crystal structure of rescued two-component self-assembling tetrahedral cage t33-31
|
|
Structure:
|
 |
Chorismate mutase. Chain: a, b, c, d, e, f, g, h, i, j, k, l. Fragment: chorismate mutase. Engineered: yes. Mutation: yes. Divalent-cation tolerance protein cuta. Chain: m, n, o, p, q, r, s, t, u, v, w, x. Fragment: divalent cation tolerance protein. Engineered: yes.
|
|
Source:
|
 |
Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579). Organism_taxid: 300852. Strain: hb8 / atcc 27634 / dsm 579. Gene: ttha0868. Expressed in: escherichia coli. Expression_system_taxid: 469008. Gene: cuta, ttha1356.
|
|
Resolution:
|
 |
|
3.40Å
|
R-factor:
|
0.192
|
R-free:
|
0.239
|
|
|
Authors:
|
 |
Y.Liu,D.Cascio,M.R.Sawaya,J.Bale,M.J.Collazo,R.Park,N.King,D.Baker, T.Yeates
|
|
Key ref:
|
 |
J.B.Bale
et al.
(2015).
Structure of a designed tetrahedral protein assembly variant engineered to have improved soluble expression.
Protein Sci,
24,
1695-1701.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
30-Apr-15
|
Release date:
|
29-Jul-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q5SJY4
(Q5SJY4_THET8) -
Chorismate mutase AroH from Thermus thermophilus (strain ATCC 27634 / DSM 579 / HB8)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
122 a.a.
117 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B, C, D, E, F, G, H, I, J, K, L:
E.C.5.4.99.5
- chorismate mutase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Phenylalanine and Tyrosine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
chorismate = prephenate
|
 |
 |
 |
 |
 |
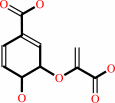 chorismate
chorismate
|
=
|
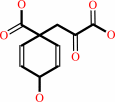 prephenate
prephenate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains M, N, O, P, Q, R, S, T, U, V, W, X:
E.C.?
|
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
24:1695-1701
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of a designed tetrahedral protein assembly variant engineered to have improved soluble expression.
|
|
J.B.Bale,
R.U.Park,
Y.Liu,
S.Gonen,
T.Gonen,
D.Cascio,
N.P.King,
T.O.Yeates,
D.Baker.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
We recently reported the development of a computational method for the design of
coassembling multicomponent protein nanomaterials. While four such materials
were validated at high-resolution by X-ray crystallography, low yield of soluble
protein prevented X-ray structure determination of a fifth designed material,
T33-09. Here we report the design and crystal structure of T33-31, a variant of
T33-09 with improved soluble yield resulting from redesign efforts focused on
mutating solvent-exposed side chains to charged amino acids. The structure is
found to match the computational design model with atomic-level accuracy,
providing further validation of the design approach and demonstrating a simple
and potentially general means of improving the yield of designed protein
nanomaterials.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| |

