 |
PDBsum entry 4zfc

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase/oxidoreductase inhibitor
|
PDB id
|
|

|

|
4zfc
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase/oxidoreductase inhibitor
|
 |
|
Title:
|
 |
Crystal structure of akr1c3 complexed with glicazide
|
|
Structure:
|
 |
Aldo-keto reductase family 1 member c3. Chain: a, b. Synonym: 17-beta-hydroxysteroid dehydrogenase type 5,17-beta-hsd 5,3- alpha-hsd type ii,brain,3-alpha-hydroxysteroid dehydrogenase type 2, 3-alpha-hsd type 2,chlordecone reductase homolog hakrb,dihydrodiol dehydrogenase 3,dd3,dihydrodiol dehydrogenase type i,ha1753,indanol dehydrogenase,prostaglandin f synthase,pgfs,testosterone 17-beta- dehydrogenase 5,trans-1,2-dihydrobenzene-1,2-diol dehydrogenase. Ec: 1.1.1.357,1.1.1.112,1.1.1.188,1.1.1.239,1.1.1.64,1.3.1.20.
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: akr1c3. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.175
|
R-free:
|
0.218
|
|
|
Authors:
|
 |
Y.Zhao,X.Zhrng,X.Hu
|
|
Key ref:
|
 |
Y.Zhao
et al.
(2015).
In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis.
Chem Biol Interact,
240,
310-315.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
21-Apr-15
|
Release date:
|
25-Nov-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P42330
(AK1C3_HUMAN) -
Aldo-keto reductase family 1 member C3 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
323 a.a.
315 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.1.188
- prostaglandin-F synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
prostaglandin F2alpha + NADP+ = prostaglandin D2 + NADPH + H+
|
 |
 |
 |
 |
 |
prostaglandin F2alpha
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 NADP(+)
NADP(+)
|
=
|
 prostaglandin D2
prostaglandin D2
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.1.1.210
- 3beta-(or 20alpha)-hydroxysteroid dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5alpha-androstane-3beta,17beta-diol + NADP+ = 17beta-hydroxy-5alpha- androstan-3-one + NADPH + H+
|
 |
 |
 |
 |
 |
5alpha-androstane-3beta,17beta-diol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
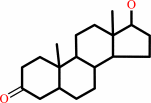 17beta-hydroxy-5alpha- androstan-3-one
17beta-hydroxy-5alpha- androstan-3-one
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.1.1.1.239
- 3alpha-(17beta)-hydroxysteroid dehydrogenase (NAD(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
testosterone + NAD+ = androst-4-ene-3,17-dione + NADH + H+
|
 |
 |
 |
 |
 |
testosterone
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
+
|
 NAD(+)
NAD(+)
|
=
|
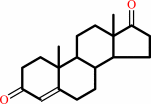 androst-4-ene-3,17-dione
androst-4-ene-3,17-dione
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
E.C.1.1.1.357
- 3alpha-hydroxysteroid 3-dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a 3alpha-hydroxysteroid + NADP+ = a 3-oxosteroid + NADPH + H+
|
|
2.
|
a 3alpha-hydroxysteroid + NAD+ = a 3-oxosteroid + NADH + H+
|
|
 |
 |
 |
 |
 |
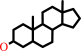 3alpha-hydroxysteroid
3alpha-hydroxysteroid
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 3-oxosteroid
3-oxosteroid
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 3alpha-hydroxysteroid
3alpha-hydroxysteroid
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
 3-oxosteroid
3-oxosteroid
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 6:
|
 |
E.C.1.1.1.53
- 3alpha(or 20beta)-hydroxysteroid dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
androstan-3alpha,17beta-diol + NAD+ = 17beta-hydroxyandrostanone + NADH + H+
|
 |
 |
 |
 |
 |
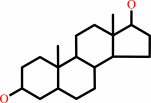 androstan-3alpha,17beta-diol
androstan-3alpha,17beta-diol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
17beta-hydroxyandrostanone
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 7:
|
 |
E.C.1.1.1.62
- 17beta-estradiol 17-dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
17beta-estradiol + NAD+ = estrone + NADH + H+
|
|
2.
|
17beta-estradiol + NADP+ = estrone + NADPH + H+
|
|
 |
 |
 |
 |
 |
17beta-estradiol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
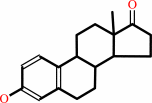 estrone
estrone
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
17beta-estradiol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 estrone
estrone
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 8:
|
 |
E.C.1.1.1.64
- testosterone 17beta-dehydrogenase (NADP(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
testosterone + NADP+ = androst-4-ene-3,17-dione + NADPH + H+
|
 |
 |
 |
 |
 |
testosterone
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 NADP(+)
NADP(+)
|
=
|
 androst-4-ene-3,17-dione
androst-4-ene-3,17-dione
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Chem Biol Interact
240:310-315
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis.
|
|
Y.Zhao,
X.Zheng,
H.Zhang,
J.Zhai,
L.Zhang,
C.Li,
K.Zeng,
Y.Chen,
Q.Li,
X.Hu.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Recent epidemiological studies show conflicting data for the first-line
anti-diabetic sulphonylureas drugs in treating cancer progression in type II
diabetes patients. How sulphonylureas promote or diminish tumor growth is not
fully understood. Here, we report that seven sulphonylureas exhibit different
in vitro inhibition towards AKR1Cs (AKR1C1, AKR1C2, AKR1C3), which are critical
steroid hormone metabolism enzymes that are related to prostate cancer, breast
cancer and endometrial diseases. Interactions of the sulphonylureas and AKR1Cs
were analyzed by X-ray crystallography.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

