 |
PDBsum entry 4tmf

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase/hydrolase inhibitor
|
PDB id
|
|

|

|
4tmf
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase/hydrolase inhibitor
|
 |
|
Title:
|
 |
Crystal structure of human cd38 in complex with hydrolysed compound jms713
|
|
Structure:
|
 |
Adp-ribosyl cyclase/cyclic adp-ribose hydrolase 1. Chain: a, b. Fragment: ecto domain. Synonym: 2'-phospho-adp-ribosyl cyclase,2'-phospho-adp-ribosyl cyclase/2'-phospho-cyclic-adp-ribose transferase,2'-phospho-cyclic- adp-ribose transferase,adp-ribosyl cyclase 1,adprc 1,cyclic adp- ribose hydrolase 1,cadpr hydrolase 1,t10. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: cd38. Expressed in: pichia pastoris. Expression_system_taxid: 4922.
|
|
Resolution:
|
 |
|
2.05Å
|
R-factor:
|
0.200
|
R-free:
|
0.227
|
|
|
Authors:
|
 |
H.Zhang,J.Swarbrick,B.Potter,Q.Hao
|
|
Key ref:
|
 |
J.M.Swarbrick
et al.
(2014).
Cyclic adenosine 5'-diphosphate ribose analogs without a "southern" ribose inhibit ADP-ribosyl cyclase-hydrolase CD38.
J Med Chem,
57,
8517-8529.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
01-Jun-14
|
Release date:
|
13-May-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P28907
(CD38_HUMAN) -
ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
300 a.a.
244 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 5 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.4.99.20
- 2'-phospho-ADP-ribosyl cyclase/2'-phospho-cyclic-ADP-ribose transferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
nicotinate + NADP+ = nicotinate-adenine dinucleotide phosphate + nicotinamide
|
 |
 |
 |
 |
 |
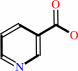 nicotinate
nicotinate
|
+
|
 NADP(+)
NADP(+)
|
=
|
nicotinate-adenine dinucleotide phosphate
|
+
|
 nicotinamide
nicotinamide
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.2.2.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.2.2.6
- ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
NAD+ + H2O = ADP-D-ribose + nicotinamide + H+
|
 |
 |
 |
 |
 |
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
ADP-D-ribose
Bound ligand (Het Group name = )
matches with 46.81% similarity
|
+
|
 nicotinamide
nicotinamide
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Med Chem
57:8517-8529
(2014)
|
|
PubMed id:
|
|
|
|

|
| |
|
Cyclic adenosine 5'-diphosphate ribose analogs without a "southern" ribose inhibit ADP-ribosyl cyclase-hydrolase CD38.
|
|
J.M.Swarbrick,
R.Graeff,
H.Zhang,
M.P.Thomas,
Q.Hao,
B.V.Potter.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cyclic adenosine 5'-diphosphate ribose (cADPR) analogs based on the cyclic
inosine 5'-diphosphate ribose (cIDPR) template were synthesized by recently
developed stereo- and regioselective N1-ribosylation. Replacing the base
N9-ribose with a butyl chain generates inhibitors of cADPR hydrolysis by the
human ADP-ribosyl cyclase CD38 catalytic domain (shCD38), illustrating the
nonessential nature of the "southern" ribose for binding. Butyl
substitution generally improves potency relative to the parent cIDPRs, and
8-amino-N9-butyl-cIDPR is comparable to the best noncovalent CD38 inhibitors to
date (IC50 = 3.3 μM). Crystallographic analysis of the
shCD38:8-amino-N9-butyl-cIDPR complex to a 2.05 Å resolution unexpectedly
reveals an N1-hydrolyzed ligand in the active site, suggesting that it is the
N6-imino form of cADPR that is hydrolyzed by CD38. While HPLC studies confirm
ligand cleavage at very high protein concentrations, they indicate that
hydrolysis does not occur under physiological concentrations. Taken together,
these analogs confirm that the "northern" ribose is critical for CD38
activity and inhibition, provide new insight into the mechanism of cADPR
hydrolysis by CD38, and may aid future inhibitor design.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

