 |
PDBsum entry 4cbb

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4cbb
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Apo form of betaine aldehyde dehydrogenase from pseudomonas aeruginosa
|
|
Structure:
|
 |
Betaine aldehyde dehydrogenase. Chain: a, b, c, d, e, f, g, h. Fragment: residues 2-490. Synonym: badh. Engineered: yes
|
|
Source:
|
 |
Pseudomonas aeruginosa. Organism_taxid: 287. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.80Å
|
R-factor:
|
0.149
|
R-free:
|
0.183
|
|
|
Authors:
|
 |
L.Gonzalez-Segura,A.G.Diaz-Sanchez,R.A.Munoz-Clares
|
|
Key ref:
|
 |
A.G.Diaz-Sanchez
et al.
The structural bases of the dual coenzyme specificity betaine aldehyde dehydrogenase from pseudomonas aerug.
To be published,
.
|
 |
|
Date:
|
 |
|
11-Oct-13
|
Release date:
|
29-Oct-14
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9HTJ1
(BETB_PSEAE) -
NAD/NADP-dependent betaine aldehyde dehydrogenase from Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
490 a.a.
489 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.2.1.8
- betaine-aldehyde dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
betaine aldehyde + NAD+ + H2O = glycine betaine + NADH + 2 H+
|
 |
 |
 |
 |
 |
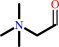 betaine aldehyde
betaine aldehyde
|
+
|
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
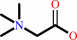 glycine betaine
glycine betaine
|
+
|
 NADH
NADH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

