 |
PDBsum entry 4bpm
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Crystal structure of a human integral membrane enzyme
|
|
Structure:
|
 |
Prostaglandin e synthase, fusion peptide. Chain: a. Fragment: gsh-binding motif, residues 10-152. Synonym: microsomal glutathione s-transferase 1-like 1, mgst1-l1, microsomal prostaglandin e synthase 1, mpges-1, p53-induced gene 12 protein, prostaglandin e2 synthase 1. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Homo sapiens, synthetic construct. Organism_taxid: 9606, 32630. Expressed in: spodoptera frugiperda. Expression_system_taxid: 7108. Expression_system_cell_line: sf9.
|
|
Resolution:
|
 |
|
2.08Å
|
R-factor:
|
0.201
|
R-free:
|
0.218
|
|
|
Authors:
|
 |
D.Li,M.Wang,V.Olieric,M.Caffrey
|
|
Key ref:
|
 |
D.Li
et al.
(2014).
Crystallizing Membrane Proteins in the Lipidic Mesophase. Experience with Human Prostaglandin E2 Synthase 1 and an Evolving Strategy.
Cryst Growth Des,
14,
2034-2047.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
27-May-13
|
Release date:
|
16-Apr-14
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.11.1.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.2.5.1.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.2.5.1.18
- glutathione transferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
RX + glutathione = an S-substituted glutathione + a halide anion + H+
|
 |
 |
 |
 |
 |
RX
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 glutathione
glutathione
|
=
|
S-substituted glutathione
|
+
|
halide anion
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
E.C.4.4.1.20
- leukotriene-C4 synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
leukotriene C4 = leukotriene A4 + glutathione
|
 |
 |
 |
 |
 |
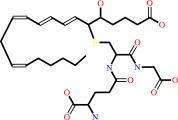 leukotriene C4
leukotriene C4
|
=
|
leukotriene A4
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 glutathione
glutathione
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 6:
|
 |
E.C.5.3.99.3
- prostaglandin-E synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
prostaglandin H2 = prostaglandin E2
|
 |
 |
 |
 |
 |
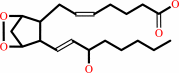 prostaglandin H2
prostaglandin H2
|
=
|
 prostaglandin E2
prostaglandin E2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Glutathione
|
 |
 |
 |
 |
 |
Glutathione
Bound ligand (Het Group name =
GSH)
corresponds exactly
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Cryst Growth Des
14:2034-2047
(2014)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallizing Membrane Proteins in the Lipidic Mesophase. Experience with Human Prostaglandin E2 Synthase 1 and an Evolving Strategy.
|
|
D.Li,
N.Howe,
A.Dukkipati,
S.T.Shah,
B.D.Bax,
C.Edge,
A.Bridges,
P.Hardwicke,
O.M.Singh,
G.Giblin,
A.Pautsch,
R.Pfau,
G.Schnapp,
M.Wang,
V.Olieric,
M.Caffrey.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

