 |
PDBsum entry 3vba
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of methanogen 3-isopropylmalate isomerase small subunit
|
|
Structure:
|
 |
Isopropylmalate/citramalate isomerase small subunit. Chain: a, b, c, d, e, f. Synonym: (r)-2-methylmalate dehydratase, (r)-citramalate dehydratase, 3-isopropylmalate dehydratase, alpha-isopropylmalate dehydratase, citraconate hydratase, isopropylmalate isomerase, ipmi, maleate hydratase, malease. Engineered: yes
|
|
Source:
|
 |
Methanocaldococcus jannaschii. Organism_taxid: 243232. Strain: dsm 2661. Gene: leud, mj1277. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.199
|
R-free:
|
0.244
|
|
|
Authors:
|
 |
K.Y.Hwang,E.H.Lee
|
|
Key ref:
|
 |
E.H.Lee
et al.
(2012).
Crystal structure of LeuD from Methanococcus jannaschii.
Biochem Biophys Res Commun,
419,
160-164.
PubMed id:
|
 |
|
Date:
|
 |
|
02-Jan-12
|
Release date:
|
14-Nov-12
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q58673
(LEUD_METJA) -
Isopropylmalate/citramalate isomerase small subunit from Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
168 a.a.
169 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.4.2.1.31
- maleate hydratase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(R)-malate = maleate + H2O
|
 |
 |
 |
 |
 |
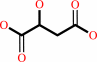 (R)-malate
(R)-malate
|
=
|
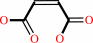 maleate
maleate
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.4.2.1.33
- 3-isopropylmalate dehydratase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R,3S)-3-isopropylmalate = (2S)-2-isopropylmalate
|
 |
 |
 |
 |
 |
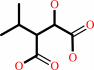 (2R,3S)-3-isopropylmalate
(2R,3S)-3-isopropylmalate
|
=
|
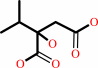 (2S)-2-isopropylmalate
(2S)-2-isopropylmalate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Iron-sulfur
|
 |
 |
 |
 |
 |
 Iron-sulfur
Iron-sulfur
|
|
 |
 |
Enzyme class 4:
|
 |
E.C.4.2.1.35
- (R)-2-methylmalate dehydratase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(3R)-citramalate = 2-methylmaleate + H2O
|
 |
 |
 |
 |
 |
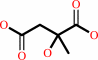 (3R)-citramalate
(3R)-citramalate
|
=
|
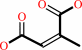 2-methylmaleate
2-methylmaleate
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochem Biophys Res Commun
419:160-164
(2012)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of LeuD from Methanococcus jannaschii.
|
|
E.H.Lee,
Y.W.Cho,
K.Y.Hwang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
3-Isopropylmalate/citramalate (IPM) isomerase catalyzes the second step in the
leucine biosynthesis pathway. IPM isomerase from Methanococcus jannaschii is a
complex protein consisting of a large (MjLeuC) and a small subunit (MjLeuD). It
has broad substrate specificity, unlike other bacterial IPM isomerases. In order
to understand the reasons for this broad substrate specificity, we determined
the crystal structure of MjLeuD at a resolution of 2.0 Å. The asymmetric unit
contained 6 molecules of LeuD, including three homodimers. The overall structure
had a β/β/α sandwich-fold consisting of 8 α-helices and 7 β-strands. The
C-terminal helix, which is important in homodimer formation, showed
conformational differences between two homodimer forms of MjLeuD. In addition,
we identified a hydrophobic residue (Val28) near the substrate recognition
region that may explain the broad substrate specificity of IPM isomerase.
Therefore, we suggest that LeuD proteins can be divided into 2 subfamilies, LeuD
subfamilies 1 and 2, which show differences in overall structure and in the
substrate recognition region.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

