
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|





 (+ 0 more)
200 a.a.
(+ 0 more)
200 a.a.
|
 |

|

|

|

|

|

|

|





 (+ 0 more)
233 a.a.
(+ 0 more)
233 a.a.
|
 |

|

|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Dioxygenase
|
 |
|
Title:
|
 |
Structure of protocatechuate 3,4-dioxygenase complexed with 3- hydroxybenzoate
|
|
Structure:
|
 |
Protocatechuate 3,4-dioxygenase alpha chain. Chain: a, b, c, d, e, f. Synonym: 3,4-pcd. Other_details: entry contains alpha/beta 6-mer. Protocatechuate 3,4-dioxygenase beta chain. Chain: m, n, o, p, q, r. Synonym: 3,4-pcd. Other_details: entry contains alpha/beta 6-mer
|
|
Source:
|
 |
Pseudomonas putida. Organism_taxid: 303. Atcc: 23975. Atcc: 23975
|
|
Biol. unit:
|
 |
24mer (from PDB file)
|
|
Resolution:
|
 |
|
|
Authors:
|
 |
N.Elango,A.M.Orville,J.D.Lipscomb,D.H.Ohlendorf
|
Key ref:

|
 |
A.M.Orville
et al.
(1997).
Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry,
36,
10039-10051.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
25-Apr-97
|
Release date:
|
29-Apr-98
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, M, B, N, C, O, D, P, E, Q, F, R:
E.C.1.13.11.3
- protocatechuate 3,4-dioxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Benzoate Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3,4-dihydroxybenzoate + O2 = 3-carboxy-cis,cis-muconate + 2 H+
|
 |
 |
 |
 |
 |
3,4-dihydroxybenzoate
Bound ligand (Het Group name = )
matches with 90.91% similarity
|
+
|
 O2
O2
|
=
|
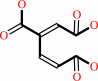 3-carboxy-cis,cis-muconate
3-carboxy-cis,cis-muconate
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
36:10039-10051
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
|
|
A.M.Orville,
N.Elango,
J.D.Lipscomb,
D.H.Ohlendorf.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Protocatechuate 3,4-dioxygenase (3,4-PCD) catalyzes the oxidative ring cleavage
of 3,4-dihydroxybenzoate to produce beta-carboxy-cis, cis-muconate. Crystal
structures of Pseudomonas putida3,4-PCD [quaternary structure of
(alphabetaFe3+)12] complexed with seven competitive inhibitors
[3-hydroxyphenylacetate (MHP), 4-hydroxyphenylacetate (PHP), 3-hydroxybenzoate
(MHB), 4-hydroxybenzoate (PHB), 3-fluoro-4-hydroxybenzoate (FHB),
3-chloro-4-hydroxybenzoate (CHB), and 3-iodo-4-hydroxybenzoate (IHB)] are
reported at 2.0-2.2 A resolution with R-factors of 0. 0.159-0.179. The
inhibitors bind in a narrow active site crevasse lined with residues that
provide a microenvironment that closely matches the chemical characteristics of
the inhibitors. This results in as little as 20% solvent-exposed surface area
for the higher-affinity inhibitors (PHB, CHB, and FHB). In uncomplexed 3,4-PCD,
the active site Fe3+ is bound at the bottom of the active site crevasse by four
endogenous ligands and a solvent molecule (Wat827). The orientations of the
endogenous ligands are relatively unperturbed in each inhibitor complex, but the
inhibitors themselves bind to or near the iron in a range of positions, all of
which perturb the position of Wat827. The three lowest-affinity inhibitors (MHP,
PHP, and IHB) yield distorted trigonal bipyramidal iron coordination geometry in
which the inhibitor C4-phenolate group displaces the solvent ligand. MHB binds
within the active site, but neither its C3-OH group nor the solvent molecule
binds to the iron. The C4-phenolate group of the three highest-affinity
inhibitors (PHB, CHB, and FHB) coordinates the Fe3+ adjacent to Wat827,
resulting in a shift in its position to yield a six-coordinate distorted
octahedral geometry. The range of inhibitor orientations may mimic the
mechanistically significant stages of substrate binding to 3, 4-PCD. The
structure of the final substrate complex is reported in the following paper
[Orville, A. M., Lipscomb, J. D., & Ohlendorf, D. H. (1997) Biochemistry 36,
10052-10066].

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.Mayilmurugan,
M.Sankaralingam,
E.Suresh,
and
M.Palaniandavar
(2010).
Novel square pyramidal iron(III) complexes of linear tetradentate bis(phenolate) ligands as structural and reactive models for intradiol-cleaving 3,4-PCD enzymes: Quinone formation vs. intradiol cleavage.
|
| |
Dalton Trans,
39,
9611-9625.
|
 |
|
|
|
|
 |
D.L.Mobley,
and
K.A.Dill
(2009).
Binding of small-molecule ligands to proteins: "what you see" is not always "what you get".
|
| |
Structure,
17,
489-498.
|
 |
|
|
|
|
 |
K.H.Kim
(2007).
Outliers in SAR and QSAR: is unusual binding mode a possible source of outliers?
|
| |
J Comput Aided Mol Des,
21,
63-86.
|
 |
|
|
|
|
 |
M.Y.Pau,
J.D.Lipscomb,
and
E.I.Solomon
(2007).
Substrate activation for O2 reactions by oxidized metal centers in biology.
|
| |
Proc Natl Acad Sci U S A,
104,
18355-18362.
|
 |
|
|
|
|
 |
M.Y.Pau,
M.I.Davis,
A.M.Orville,
J.D.Lipscomb,
and
E.I.Solomon
(2007).
Spectroscopic and electronic structure study of the enzyme-substrate complex of intradiol dioxygenases: substrate activation by a high-spin ferric non-heme iron site.
|
| |
J Am Chem Soc,
129,
1944-1958.
|
 |
|
|
|
|
 |
M.Ferraroni,
J.Seifert,
V.M.Travkin,
M.Thiel,
S.Kaschabek,
A.Scozzafava,
L.Golovleva,
M.Schlömann,
and
F.Briganti
(2005).
Crystal structure of the hydroxyquinol 1,2-dioxygenase from Nocardioides simplex 3E, a key enzyme involved in polychlorinated aromatics biodegradation.
|
| |
J Biol Chem,
280,
21144-21154.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.L.Neidig,
and
E.I.Solomon
(2005).
Structure-function correlations in oxygen activating non-heme iron enzymes.
|
| |
Chem Commun (Camb),
(),
5843-5863.
|
 |
|
|
|
|
 |
C.K.Brown,
M.W.Vetting,
C.A.Earhart,
and
D.H.Ohlendorf
(2004).
Biophysical analyses of designed and selected mutants of protocatechuate 3,4-dioxygenase1.
|
| |
Annu Rev Microbiol,
58,
555-585.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Ferraroni,
I.P.Solyanikova,
M.P.Kolomytseva,
A.Scozzafava,
L.Golovleva,
and
F.Briganti
(2004).
Crystal structure of 4-chlorocatechol 1,2-dioxygenase from the chlorophenol-utilizing gram-positive Rhodococcus opacus 1CP.
|
| |
J Biol Chem,
279,
27646-27655.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Potrawfke,
J.Armengaud,
and
R.M.Wittich
(2001).
Chlorocatechols substituted at positions 4 and 5 are substrates of the broad-spectrum chlorocatechol 1,2-dioxygenase of Pseudomonas chlororaphis RW71.
|
| |
J Bacteriol,
183,
997.
|
 |
|
|
|
|
 |
M.W.Vetting,
D.A.D'Argenio,
L.N.Ornston,
and
D.H.Ohlendorf
(2000).
Structure of Acinetobacter strain ADP1 protocatechuate 3, 4-dioxygenase at 2.2 A resolution: implications for the mechanism of an intradiol dioxygenase.
|
| |
Biochemistry,
39,
7943-7955.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.W.Vetting,
and
D.H.Ohlendorf
(2000).
The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
|
| |
Structure,
8,
429-440.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.J.Schofield,
and
Z.Zhang
(1999).
Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes.
|
| |
Curr Opin Struct Biol,
9,
722-731.
|
 |
|
|
|
|
 |
D.A.D'Argenio,
M.W.Vetting,
D.H.Ohlendorf,
and
L.N.Ornston
(1999).
Substitution, insertion, deletion, suppression, and altered substrate specificity in functional protocatechuate 3,4-dioxygenases.
|
| |
J Bacteriol,
181,
6478-6487.
|
 |
|
|
|
|
 |
F.X.Gomis-Rüth,
V.Companys,
Y.Qian,
L.D.Fricker,
J.Vendrell,
F.X.Avilés,
and
M.Coll
(1999).
Crystal structure of avian carboxypeptidase D domain II: a prototype for the regulatory metallocarboxypeptidase subfamily.
|
| |
EMBO J,
18,
5817-5826.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Sugimoto,
T.Senda,
H.Aoshima,
E.Masai,
M.Fukuda,
and
Y.Mitsui
(1999).
Crystal structure of an aromatic ring opening dioxygenase LigAB, a protocatechuate 4,5-dioxygenase, under aerobic conditions.
|
| |
Structure,
7,
953-965.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.K.Lee,
and
J.D.Lipscomb
(1999).
Oxygen activation catalyzed by methane monooxygenase hydroxylase component: proton delivery during the O-O bond cleavage steps.
|
| |
Biochemistry,
38,
4423-4432.
|
 |
|
|
|
|
 |
A.J.Thomson,
and
H.B.Gray
(1998).
Bio-inorganic chemistry
|
| |
Curr Opin Chem Biol,
2,
155-158.
|
 |
|
|
|
|
 |
R.W.Frazee,
A.M.Orville,
K.B.Dolbeare,
H.Yu,
D.H.Ohlendorf,
and
J.D.Lipscomb
(1998).
The axial tyrosinate Fe3+ ligand in protocatechuate 3,4-dioxygenase influences substrate binding and product release: evidence for new reaction cycle intermediates.
|
| |
Biochemistry,
37,
2131-2144.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.J.Lange,
and
L.Que
(1998).
Oxygen activating nonheme iron enzymes.
|
| |
Curr Opin Chem Biol,
2,
159-172.
|
 |
|
|
|
|
 |
A.M.Orville,
J.D.Lipscomb,
and
D.H.Ohlendorf
(1997).
Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
|
| |
Biochemistry,
36,
10052-10066.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.M.Orville,
and
J.D.Lipscomb
(1997).
Cyanide and nitric oxide binding to reduced protocatechuate 3,4-dioxygenase: insight into the basis for order-dependent ligand binding by intradiol catecholic dioxygenases.
|
| |
Biochemistry,
36,
14044-14055.
|
 |
|
|
|
|
 |
T.E.Elgren,
A.M.Orville,
K.A.Kelly,
J.D.Lipscomb,
D.H.Ohlendorf,
and
L.Que
(1997).
Crystal structure and resonance Raman studies of protocatechuate 3,4-dioxygenase complexed with 3,4-dihydroxyphenylacetate.
|
| |
Biochemistry,
36,
11504-11513.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

