 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.112
- indanol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
(S)-indan-1-ol + NAD+ = indan-1-one + NADH + H+
|
|
2.
|
(S)-indan-1-ol + NADP+ = indan-1-one + NADPH + H+
|
|
 |
 |
 |
 |
 |
(S)-indan-1-ol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
indan-1-one
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
(S)-indan-1-ol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
indan-1-one
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.1.149
- 20alpha-hydroxysteroid dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
(17R,20S)-17,20-dihydroxypregn-4-en-3-one + NAD+ = 17alpha- hydroxyprogesterone + NADH + H+
|
|
2.
|
(17R,20S)-17,20-dihydroxypregn-4-en-3-one + NADP+ = 17alpha- hydroxyprogesterone + NADPH + H+
|
|
 |
 |
 |
 |
 |
(17R,20S)-17,20-dihydroxypregn-4-en-3-one
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
17alpha- hydroxyprogesterone
Bound ligand (Het Group name = )
matches with 51.85% similarity
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
(17R,20S)-17,20-dihydroxypregn-4-en-3-one
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
17alpha- hydroxyprogesterone
Bound ligand (Het Group name = )
matches with 51.85% similarity
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.1.1.209
- 3(or 17)alpha-hydroxysteroid dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
androsterone + NADP+ = 5alpha-androstan-3,17-dione + NADPH + H+
|
|
2.
|
androsterone + NAD+ = 5alpha-androstan-3,17-dione + NADH + H+
|
|
 |
 |
 |
 |
 |
androsterone
Bound ligand (Het Group name = )
matches with 58.33% similarity
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
5alpha-androstan-3,17-dione
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
androsterone
Bound ligand (Het Group name = )
matches with 58.33% similarity
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
5alpha-androstan-3,17-dione
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.1.1.1.210
- 3beta-(or 20alpha)-hydroxysteroid dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5alpha-androstane-3beta,17beta-diol + NADP+ = 17beta-hydroxy-5alpha- androstan-3-one + NADPH + H+
|
 |
 |
 |
 |
 |
5alpha-androstane-3beta,17beta-diol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
17beta-hydroxy-5alpha- androstan-3-one
Bound ligand (Het Group name = )
matches with 58.33% similarity
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
E.C.1.1.1.357
- 3alpha-hydroxysteroid 3-dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a 3alpha-hydroxysteroid + NADP+ = a 3-oxosteroid + NADPH + H+
|
|
2.
|
a 3alpha-hydroxysteroid + NAD+ = a 3-oxosteroid + NADH + H+
|
|
 |
 |
 |
 |
 |
3alpha-hydroxysteroid
Bound ligand (Het Group name = )
matches with 60.87% similarity
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 3-oxosteroid
3-oxosteroid
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
3alpha-hydroxysteroid
Bound ligand (Het Group name = )
matches with 60.87% similarity
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
 3-oxosteroid
3-oxosteroid
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 6:
|
 |
E.C.1.1.1.51
- 3(or 17)beta-hydroxysteroid dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
testosterone + NAD+ = androst-4-ene-3,17-dione + NADH + H+
|
|
2.
|
testosterone + NADP+ = androst-4-ene-3,17-dione + NADPH + H+
|
|
 |
 |
 |
 |
 |
testosterone
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 58.33% similarity
|
=
|
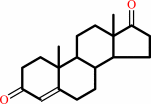 androst-4-ene-3,17-dione
androst-4-ene-3,17-dione
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
testosterone
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
NADP(+)
Bound ligand (Het Group name = )
matches with 58.33% similarity
|
=
|
 androst-4-ene-3,17-dione
androst-4-ene-3,17-dione
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 7:
|
 |
E.C.1.1.1.53
- 3alpha(or 20beta)-hydroxysteroid dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
androstan-3alpha,17beta-diol + NAD+ = 17beta-hydroxyandrostanone + NADH + H+
|
 |
 |
 |
 |
 |
androstan-3alpha,17beta-diol
Bound ligand (Het Group name = )
matches with 58.33% similarity
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
17beta-hydroxyandrostanone
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 8:
|
 |
E.C.1.1.1.62
- 17beta-estradiol 17-dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
17beta-estradiol + NAD+ = estrone + NADH + H+
|
|
2.
|
17beta-estradiol + NADP+ = estrone + NADPH + H+
|
|
 |
 |
 |
 |
 |
17beta-estradiol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
estrone
Bound ligand (Het Group name = )
matches with 60.87% similarity
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
17beta-estradiol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
estrone
Bound ligand (Het Group name = )
matches with 60.87% similarity
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 9:
|
 |
E.C.1.3.1.20
- trans-1,2-dihydrobenzene-1,2-diol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
(1R,2R)-1,2-dihydrobenzene-1,2-diol + NADP+ = catechol + NADPH + H+
|
|
2.
|
(1S,2S)-1,2-dihydrobenzene-1,2-diol + NADP+ = catechol + NADPH + H+
|
|
 |
 |
 |
 |
 |
(1R,2R)-1,2-dihydrobenzene-1,2-diol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
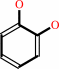 catechol
catechol
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
(1S,2S)-1,2-dihydrobenzene-1,2-diol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 catechol
catechol
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |

