 |
PDBsum entry 3ltp

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with inhibitor bmp
|
|
Structure:
|
 |
Orotidine 5'-phosphate decarboxylase. Chain: a, b. Synonym: omp decarboxylase, ompdcase, ompdecase. Engineered: yes
|
|
Source:
|
 |
Methanothermobacter thermautotrophicus. Organism_taxid: 187420. Strain: delta h. Gene: pyrf, mth_129. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.40Å
|
R-factor:
|
0.184
|
R-free:
|
0.195
|
|
|
Authors:
|
 |
A.A.Fedorov,E.V.Fedorov,B.M.Wood,J.A.Gerlt,S.C.Almo
|
|
Key ref:
|
 |
B.M.Wood
et al.
(2010).
Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry,
49,
3514-3516.
PubMed id:
|
 |
|
Date:
|
 |
|
16-Feb-10
|
Release date:
|
16-Jun-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O26232
(PYRF_METTH) -
Orotidine 5'-phosphate decarboxylase from Methanothermobacter thermautotrophicus (strain ATCC 29096 / DSM 1053 / JCM 10044 / NBRC 100330 / Delta H)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
228 a.a.
218 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.23
- orotidine-5'-phosphate decarboxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pyrimidine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
orotidine 5'-phosphate + H+ = UMP + CO2
|
 |
 |
 |
 |
 |
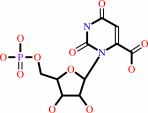 orotidine 5'-phosphate
orotidine 5'-phosphate
|
+
|
H(+)
|
=
|
 UMP
UMP
|
+
|
CO2
Bound ligand (Het Group name = )
matches with 95.45% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochemistry
49:3514-3516
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
|
|
B.M.Wood,
T.L.Amyes,
A.A.Fedorov,
E.V.Fedorov,
A.Shabila,
S.C.Almo,
J.P.Richard,
J.A.Gerlt.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structural factors responsible for the extraordinary rate enhancement (
approximately 10(17)) of the reaction catalyzed by orotidine 5'-monophosphate
decarboxylase (OMPDC) have not been defined. Catalysis requires a conformational
change that closes an active site loop and "clamps" the orotate base proximal to
hydrogen-bonded networks that destabilize the substrate and stabilize the
intermediate. In the OMPDC from Methanobacter thermoautotrophicus, a "remote"
structurally conserved cluster of hydrophobic residues that includes Val 182 in
the active site loop is assembled in the closed, catalytically active
conformation. Substitution of these residues with Ala decreases k(cat)/K(m) with
a minimal effect on k(cat), providing evidence that the cluster stabilizes the
closed conformation. The intrinsic binding energies of the 5'-phosphate group of
orotidine 5'-monophosphate for the mutant enzymes are similar to that for the
wild type, supporting this conclusion.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

