 |
PDBsum entry 3djc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of pantothenate kinase from legionella pneumophila
|
|
Structure:
|
 |
Type iii pantothenate kinase. Chain: a, b, c, d, e, f, g, h, i, j, k, l. Synonym: pantothenic acid kinase, pank-iii. Engineered: yes
|
|
Source:
|
 |
Legionella pneumophila subsp. Pneumophila str. Philadelphia 1. Organism_taxid: 272624. Gene: coax, lpg0911
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.261
|
R-free:
|
0.313
|
|
|
Authors:
|
 |
Y.Patskovsky,J.B.Bonanno,R.Romero,M.Dickey,C.Logan,S.Wasserman, M.Maletic,J.Koss,J.M.Sauder,S.K.Burley,S.C.Almo,New York Sgx Research Center For Structural Genomics (Nysgxrc)
|
|
Key ref:
|
 |
Y.Patskovsky
et al.
Crystal structure of pantothenate kinase from legionella pneumophila.
To be published,
.
|
 |
|
Date:
|
 |
|
23-Jun-08
|
Release date:
|
15-Jul-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q5ZX22
(COAX_LEGPH) -
Type III pantothenate kinase from Legionella pneumophila subsp. pneumophila (strain Philadelphia 1 / ATCC 33152 / DSM 7513)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
256 a.a.
255 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.33
- pantothenate kinase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Coenzyme A Biosynthesis (late stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(R)-pantothenate + ATP = (R)-4'-phosphopantothenate + ADP + H+
|
 |
 |
 |
 |
 |
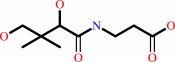 (R)-pantothenate
(R)-pantothenate
|
+
|
 ATP
ATP
|
=
|
 (R)-4'-phosphopantothenate
(R)-4'-phosphopantothenate
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

