 |
PDBsum entry 3d0f

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.4.99.28
- peptidoglycan glycosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
[GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)](n)- di-trans,octa-cis-undecaprenyl diphosphate + beta-D-GlcNAc-(1->4)- Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-di-trans,octa- cis-undecaprenyl diphosphate = [GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D- Glu-L-Lys-D-Ala-D-Ala)](n+1)-di-trans,octa-cis-undecaprenyl diphosphate + di-trans,octa-cis-undecaprenyl diphosphate + H+
|
 |
 |
 |
 |
 |
[GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)](n)- di-trans,octa-cis-undecaprenyl diphosphate
|
+
|
beta-D-GlcNAc-(1->4)- Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-di-trans,octa- cis-undecaprenyl diphosphate
|
=
|
[GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D- Glu-L-Lys-D-Ala-D-Ala)](n+1)-di-trans,octa-cis-undecaprenyl diphosphate
|
+
|
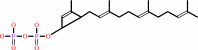 di-trans,octa-cis-undecaprenyl diphosphate
di-trans,octa-cis-undecaprenyl diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.4.16.4
- serine-type D-Ala-D-Ala carboxypeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
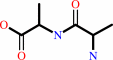
D-alanyl-D-alanine + H2O = 2 D-alanine
|
 |
 |
 |
 |
 |
 [GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)](n)- di-trans,octa-cis-undecaprenyl diphosphate
[GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)](n)- di-trans,octa-cis-undecaprenyl diphosphate
|
+
|
beta-D-GlcNAc-(1->4)- Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-di-trans,octa- cis-undecaprenyl diphosphate
|
=
|
 2
×
[GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D- Glu-L-Lys-D-Ala-D-Ala)](n+1)-di-trans,octa-cis-undecaprenyl diphosphate
2
×
[GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D- Glu-L-Lys-D-Ala-D-Ala)](n+1)-di-trans,octa-cis-undecaprenyl diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

