 |
PDBsum entry 3b6h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Crystal structure of human prostacyclin synthase in complex with inhibitor minoxidil
|
|
Structure:
|
 |
Prostacyclin synthase. Chain: a, b. Fragment: unp residues 18-500. Synonym: cytochrome p450 8a1, prostaglandin i2 synthase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: ptgis, cyp8, cyp8a1. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.62Å
|
R-factor:
|
0.204
|
R-free:
|
0.228
|
|
|
Authors:
|
 |
Y.-C.Li,C.-W.Chiang,H.-C.Yeh,P.-Y.Hsu,F.G.Whitby,L.-H.Wang,N.-L.Chan
|
Key ref:

|
 |
Y.C.Li
et al.
(2008).
Structures of Prostacyclin Synthase and Its Complexes with Substrate Analog and Inhibitor Reveal a Ligand-specific Heme Conformation Change.
J Biol Chem,
283,
2917-2926.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
29-Oct-07
|
Release date:
|
20-Nov-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q16647
(PTGIS_HUMAN) -
Prostacyclin synthase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
500 a.a.
469 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.4.2.1.152
- hydroperoxy icosatetraenoate dehydratase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a hydroperoxyeicosatetraenoate = an oxoeicosatetraenoate + H2O
|
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe(2+)
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.5.3.99.4
- prostaglandin-I synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
prostaglandin H2 = prostaglandin I2
|
 |
 |
 |
 |
 |
prostaglandin H2
Bound ligand (Het Group name = )
matches with 51.11% similarity
|
=
|
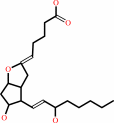 prostaglandin I2
prostaglandin I2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme-thiolate
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
283:2917-2926
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of Prostacyclin Synthase and Its Complexes with Substrate Analog and Inhibitor Reveal a Ligand-specific Heme Conformation Change.
|
|
Y.C.Li,
C.W.Chiang,
H.C.Yeh,
P.Y.Hsu,
F.G.Whitby,
L.H.Wang,
N.L.Chan.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Prostacyclin synthase (PGIS) is a cytochrome P450 (P450) enzyme that catalyzes
production of prostacyclin from prostaglandin H(2). PGIS is unusual in that it
catalyzes an isomerization rather than a monooxygenation, which is typical of
P450 enzymes. To understand the structural basis for prostacyclin biosynthesis
in greater detail, we have determined the crystal structures of ligand-free,
inhibitor (minoxidil)-bound and substrate analog U51605-bound PGIS. These
structures demonstrate a stereo-specific substrate binding and suggest features
of the enzyme that facilitate isomerization. Unlike most microsomal P450s, where
large substrate-induced conformational changes take place at the distal side of
the heme, conformational changes in PGIS are observed at the proximal side and
in the heme itself. The conserved and extensive heme propionate-protein
interactions seen in all other P450s, which are largely absent in the
ligand-free PGIS, are recovered upon U51605 binding accompanied by water
exclusion from the active site. In contrast, when minoxidil binds, the
propionate-protein interactions are not recovered and water molecules are
largely retained. These findings suggest that PGIS represents a divergent
evolution of the P450 family, in which a heme barrier has evolved to ensure
strict binding specificity for prostaglandin H(2), leading to a radical-mediated
isomerization with high product fidelity. The U51605-bound structure also
provides a view of the substrate entrance and product exit channels.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIGURE 1. Proposed mechanism for PGI[2] biosynthesis. A,
chemical structures of substrate PGH[2], substrate analog
U51605, and inhibitor minoxidil. B, a proposed mechanism for the
reaction catalyzed by PGIS (R1, CH[2]-(CH[2])[2]-COOH; R2,
CH-CHOH-(CH[2])[4]-CH[3]) (6).
|
 |
Figure 5.
FIGURE 5. The minoxidil-bound hPGIS. A, a F[o] - F[c] omit
map contoured at 2.7  shows unbiased electron
density for minoxidil. B, structural changes around the active
site, Cys ligand loop, and B' helix upon minoxidil binding. The
ligand-free (blue) and minoxidil-bound (gold) structures were
superimposed over all equivalent C shows unbiased electron
density for minoxidil. B, structural changes around the active
site, Cys ligand loop, and B' helix upon minoxidil binding. The
ligand-free (blue) and minoxidil-bound (gold) structures were
superimposed over all equivalent C  atom pairs. The
minoxidil-induced stacking between Phe^96 and His^438 is
indicated by the dashed lines. atom pairs. The
minoxidil-induced stacking between Phe^96 and His^438 is
indicated by the dashed lines.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by the ASBMB:
J Biol Chem
(2008,
283,
2917-2926)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.V.Goldstone,
A.G.McArthur,
A.Kubota,
J.Zanette,
T.Parente,
M.E.Jönsson,
D.R.Nelson,
and
J.J.Stegeman
(2010).
Identification and developmental expression of the full complement of Cytochrome P450 genes in Zebrafish.
|
| |
BMC Genomics,
11,
643.
|
 |
|
|
|
|
 |
P.Paragi-Vedanthi,
and
M.Doble
(2010).
Comparison of PGH2 binding site in prostaglandin synthases.
|
| |
BMC Bioinformatics,
11,
S51.
|
 |
|
|
|
|
 |
T.C.Pochapsky,
S.Kazanis,
and
M.Dang
(2010).
Conformational plasticity and structure/function relationships in cytochromes P450.
|
| |
Antioxid Redox Signal,
13,
1273-1296.
|
 |
|
|
|
|
 |
T.K.Yanai,
and
S.Mori
(2009).
Density functional studies on isomerization of prostaglandin H2 to prostacyclin catalyzed by cytochrome P450.
|
| |
Chemistry,
15,
4464-4473.
|
 |
|
|
|
|
 |
L.Li,
Z.Chang,
Z.Pan,
Z.Q.Fu,
and
X.Wang
(2008).
Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase.
|
| |
Proc Natl Acad Sci U S A,
105,
13883-13888.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.K.Yanai,
and
S.Mori
(2008).
Density functional studies on thromboxane biosynthesis: mechanism and role of the heme-thiolate system.
|
| |
Chem Asian J,
3,
1900-1911.
|
 |
|
|
|
|
 |
Z.Chang,
L.Li,
Z.Pan,
and
X.Wang
(2008).
Crystallization and preliminary X-ray analysis of allene oxide synthase, cytochrome P450 CYP74A2, from Parthenium argentatum.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
668-670.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

