 |
PDBsum entry 3fsl

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of tyrosine aminotransferase tripple mutant (p181q, r183g,a321k) from escherichia coli at 2.35 a resolution
|
|
Structure:
|
 |
Aromatic-amino-acid aminotransferase. Chain: a, b, c, d, e, f. Synonym: tyrosine aminotransferase, aroat, arat. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Escherichia coli k-12. Organism_taxid: 83333. Strain: bl21. Gene: b4054, jw4014, tat, tyrb. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.35Å
|
R-factor:
|
0.215
|
R-free:
|
0.260
|
|
|
Authors:
|
 |
V.N.Malashkevich,B.Ng,J.F.Kirsch
|
|
Key ref:
|
 |
V.N.Malashkevich
et al.
Crystal structure of tyrosine aminotransferase tripple mutant (p181q,R183g,A321k) from escherichia coli at 2.35 a resolution.
To be published,
.
|
 |
|
Date:
|
 |
|
09-Jan-09
|
Release date:
|
27-Jan-09
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P04693
(TYRB_ECOLI) -
Aromatic-amino-acid aminotransferase from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
397 a.a.
397 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 3 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.6.1.107
- beta-methylphenylalanine transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(3S)-3-methyl-L-phenylalanine + 2-oxoglutarate = (3S)-2-oxo-3- phenylbutanoate + L-glutamate
|
 |
 |
 |
 |
 |
(3S)-3-methyl-L-phenylalanine
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
(3S)-2-oxo-3- phenylbutanoate
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLR)
matches with 93.75% similarity
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.2.6.1.57
- aromatic-amino-acid transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an aromatic L-alpha-amino acid + 2-oxoglutarate = an aromatic oxo-acid + L-glutamate
|
 |
 |
 |
 |
 |
aromatic L-alpha-amino acid
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
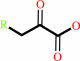 aromatic oxo-acid
aromatic oxo-acid
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLR)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

