 |
PDBsum entry 3epm

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Biosynthetic protein
|
PDB id
|
|

|

|
3epm
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Biosynthetic protein
|
 |
|
Title:
|
 |
Crystal structure of caulobacter crescentus thic
|
|
Structure:
|
 |
Thiamine biosynthesis protein thic. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Caulobacter crescentus. Organism_taxid: 155892. Gene: thic. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.79Å
|
R-factor:
|
0.185
|
R-free:
|
0.244
|
|
|
Authors:
|
 |
S.Li,A.Chatterjee,Y.Zhang,T.L.Grove,M.Lee,C.Krebs,S.J.Booker, T.P.Begley,S.E.Ealick
|
Key ref:

|
 |
A.Chatterjee
et al.
(2008).
Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily.
Nat Chem Biol,
4,
758-765.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
29-Sep-08
|
Release date:
|
28-Oct-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9A6Q5
(THIC_CAUVC) -
Phosphomethylpyrimidine synthase from Caulobacter vibrioides (strain ATCC 19089 / CIP 103742 / CB 15)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
612 a.a.
515 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.99.17
- phosphomethylpyrimidine synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5-amino-1-(5-phospho-beta-D-ribosyl)imidazole + S-adenosyl-L-methionine = 4-amino-2-methyl-5-(phosphooxymethyl)pyrimidine + CO + 5'-deoxyadenosine + formate + L-methionine + 3 H+
|
 |
 |
 |
 |
 |
5-amino-1-(5-phospho-beta-D-ribosyl)imidazole
|
+
|
 S-adenosyl-L-methionine
S-adenosyl-L-methionine
|
=
|
4-amino-2-methyl-5-(phosphooxymethyl)pyrimidine
|
+
|
CO
Bound ligand (Het Group name = )
matches with 55.56% similarity
|
+
|
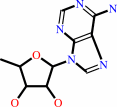 5'-deoxyadenosine
5'-deoxyadenosine
|
+
|
 formate
formate
|
+
|
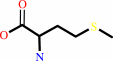 L-methionine
L-methionine
|
+
|
3
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Iron-sulfur
|
 |
 |
 |
 |
 |
 Iron-sulfur
Iron-sulfur
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Chem Biol
4:758-765
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily.
|
|
A.Chatterjee,
Y.Li,
Y.Zhang,
T.L.Grove,
M.Lee,
C.Krebs,
S.J.Booker,
T.P.Begley,
S.E.Ealick.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
4-Amino-5-hydroxymethyl-2-methylpyrimidine phosphate (HMP-P) synthase catalyzes
a complex rearrangement of 5-aminoimidazole ribonucleotide (AIR) to form HMP-P,
the pyrimidine moiety of thiamine phosphate. We determined the three-dimensional
structures of HMP-P synthase and its complexes with the product HMP-P and a
substrate analog imidazole ribotide. The structure of HMP-P synthase reveals a
homodimer in which each protomer comprises three domains: an N-terminal domain
with a novel fold, a central (betaalpha)(8) barrel and a disordered C-terminal
domain that contains a conserved CX(2)CX(4)C motif, which is suggestive of a
[4Fe-4S] cluster. Biochemical studies have confirmed that HMP-P synthase is iron
sulfur cluster-dependent, that it is a new member of the radical SAM superfamily
and that HMP-P and 5'-deoxyadenosine are products of the reaction. Mössbauer
and EPR spectroscopy confirm the presence of one [4Fe-4S] cluster. Structural
comparisons reveal that HMP-P synthase is homologous to a group of
adenosylcobalamin radical enzymes. This similarity supports an evolutionary
relationship between these two superfamilies.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
(a) Overall bacterial pathway. AIR (12) is converted to HMP-P
(15) by HMP-P synthase (ThiC), which is phosphorylated by ThiD
to give HMP pyrophosphate (3). The thiazole moiety (2) is
biosynthesized from DXP (6), cysteine (8) and dehydroglycine
(34). The dehydroglycine is generated from glycine (ThiO) in B.
subtilis and from tyrosine (ThiH) in E. coli. The pyrimidine and
thiazole are coupled by ThiE to give thiamine phosphate (4), and
ThiL catalyzes the final phosphorylation. (b) Conversion of AIR
to the thiamine pyrimidine in bacteria and plants. The color
coding indicates the source of nonhydrogen atoms in HMP-P as
demonstrated by labeling studies. (c) Biosynthesis of thiamine
pyrimidine in fungi. In fungi the pyrimidine moiety is derived
from histidine (13) and pyridoxal 5'-phosphate (14) using a
single enzyme, THI5p. The color coding indicates the source of
nonhydrogen atoms. (d) The HMP-P synthase reactions. When
iron-sulfur cluster–loaded HMP-P synthase is reduced with
dithionite, it reduces SAM (16) to generate methionine (28) and
the 5'-deoxyadenosyl (5-dAdo) radical (17), which is required by
HMP-P synthase to convert AIR to HMP-P.
|
 |
Figure 4.
(a) The HMP-P synthase homodimer. The protomer consists of
three domains. The N-terminal domains are colored in shades of
blue, the (   )[8]
core domains are colored in shades of green and the C-terminal
domains are colored in shades of red. HMP-P is shown as a
ball-and-stick model. The final 66 amino acids are disordered;
however, the final ordered residues, which immediately precede a
conserved CX[2]CX[4]C motif, extend into the active site of the
adjacent protomer. The C-terminal tail is anchored to the
adjacent protomer by a three-helix bundle motif located at the
beginning of the C-terminal domain. (b) Stereoview of the HMP-P
synthase active site with modeled SAM and the [4Fe-4S] cluster.
The atoms are color coded by atom type (green = C, blue = N, red
= O, yellow = S and orange = Fe). The substrate analog IMR 22
from the crystal structure is shown. Residues Cys561, Cys564 and
Cys569, SAM and the [4Fe-4S] cluster were modeled using biotin
synthase as a guide. Hydrogen bonds are indicated by dotted
lines. (c) Superposition of the ( )[8]
core domains are colored in shades of green and the C-terminal
domains are colored in shades of red. HMP-P is shown as a
ball-and-stick model. The final 66 amino acids are disordered;
however, the final ordered residues, which immediately precede a
conserved CX[2]CX[4]C motif, extend into the active site of the
adjacent protomer. The C-terminal tail is anchored to the
adjacent protomer by a three-helix bundle motif located at the
beginning of the C-terminal domain. (b) Stereoview of the HMP-P
synthase active site with modeled SAM and the [4Fe-4S] cluster.
The atoms are color coded by atom type (green = C, blue = N, red
= O, yellow = S and orange = Fe). The substrate analog IMR 22
from the crystal structure is shown. Residues Cys561, Cys564 and
Cys569, SAM and the [4Fe-4S] cluster were modeled using biotin
synthase as a guide. Hydrogen bonds are indicated by dotted
lines. (c) Superposition of the (   )[8]
domains from HMP-P synthase and biotin synthase (PDB ID 1R3O).
HMP-P synthase is shown in blue, and biotin synthase is shown in
silver. The [4Fe-4S] cluster and SAM from biotin synthase are
shown as ball-and-stick models. )[8]
domains from HMP-P synthase and biotin synthase (PDB ID 1R3O).
HMP-P synthase is shown in blue, and biotin synthase is shown in
silver. The [4Fe-4S] cluster and SAM from biotin synthase are
shown as ball-and-stick models.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
Nat Chem Biol
(2008,
4,
758-765)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.J.Arcinas,
and
S.J.Booker
(2011).
Enzymology: Radical break-up, blissful make-up.
|
| |
Nat Chem Biol,
7,
133-134.
|
 |
|
|
|
|
 |
P.L.Roach
(2011).
Radicals from S-adenosylmethionine and their application to biosynthesis.
|
| |
Curr Opin Chem Biol,
15,
267-275.
|
 |
|
|
|
|
 |
Q.Zhang,
Y.Li,
D.Chen,
Y.Yu,
L.Duan,
B.Shen,
and
W.Liu
(2011).
Radical-mediated enzymatic carbon chain fragmentation-recombination.
|
| |
Nat Chem Biol,
7,
154-160.
|
 |
|
|
|
|
 |
E.N.Marsh,
D.P.Patterson,
and
L.Li
(2010).
Adenosyl radical: reagent and catalyst in enzyme reactions.
|
| |
Chembiochem,
11,
604-621.
|
 |
|
|
|
|
 |
F.Yan,
J.M.LaMarre,
R.Röhrich,
J.Wiesner,
H.Jomaa,
A.S.Mankin,
and
D.G.Fujimori
(2010).
RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA.
|
| |
J Am Chem Soc,
132,
3953-3964.
|
 |
|
|
|
|
 |
M.J.Koenigsknecht,
and
D.M.Downs
(2010).
Thiamine biosynthesis can be used to dissect metabolic integration.
|
| |
Trends Microbiol,
18,
240-247.
|
 |
|
|
|
|
 |
M.R.Challand,
F.T.Martins,
and
P.L.Roach
(2010).
Catalytic activity of the anaerobic tyrosine lyase required for thiamine biosynthesis in Escherichia coli.
|
| |
J Biol Chem,
285,
5240-5248.
|
 |
|
|
|
|
 |
R.K.Thauer,
A.K.Kaster,
M.Goenrich,
M.Schick,
T.Hiromoto,
and
S.Shima
(2010).
Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage.
|
| |
Annu Rev Biochem,
79,
507-536.
|
 |
|
|
|
|
 |
S.E.McGlynn,
E.S.Boyd,
E.M.Shepard,
R.K.Lange,
R.Gerlach,
J.B.Broderick,
and
J.W.Peters
(2010).
Identification and characterization of a novel member of the radical AdoMet enzyme superfamily and implications for the biosynthesis of the Hmd hydrogenase active site cofactor.
|
| |
J Bacteriol,
192,
595-598.
|
 |
|
|
|
|
 |
S.J.Booker,
and
T.L.Grove
(2010).
Mechanistic and functional versatility of radical SAM enzymes.
|
| |
F1000 Biol Rep,
2,
52.
|
 |
|
|
|
|
 |
Y.Zhang,
X.Zhu,
A.T.Torelli,
M.Lee,
B.Dzikovski,
R.M.Koralewski,
E.Wang,
J.Freed,
C.Krebs,
S.E.Ealick,
and
H.Lin
(2010).
Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
|
| |
Nature,
465,
891-896.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.T.Jurgenson,
T.P.Begley,
and
S.E.Ealick
(2009).
The structural and biochemical foundations of thiamin biosynthesis.
|
| |
Annu Rev Biochem,
78,
569-603.
|
 |
|
|
|
|
 |
E.McDonald
(2009).
Vitamins and cofactors: highlights of ESBOC 2009.
|
| |
Nat Chem Biol,
5,
530-533.
|
 |
|
|
|
|
 |
K.H.Lee,
L.Saleh,
B.P.Anton,
C.L.Madinger,
J.S.Benner,
D.F.Iwig,
R.J.Roberts,
C.Krebs,
and
S.J.Booker
(2009).
Characterization of RimO, a new member of the methylthiotransferase subclass of the radical SAM superfamily.
|
| |
Biochemistry,
48,
10162-10174.
|
 |
|
|
|
|
 |
N.C.Martinez-Gomez,
R.R.Poyner,
S.O.Mansoorabadi,
G.H.Reed,
and
D.M.Downs
(2009).
Reaction of AdoMet with ThiC generates a backbone free radical.
|
| |
Biochemistry,
48,
217-219.
|
 |
|
|
|
|
 |
P.H.Szu,
M.W.Ruszczycky,
S.H.Choi,
F.Yan,
and
H.W.Liu
(2009).
Characterization and mechanistic studies of DesII: a radical S-adenosyl-L-methionine enzyme involved in the biosynthesis of TDP-D-desosamine.
|
| |
J Am Chem Soc,
131,
14030-14042.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

