 |
PDBsum entry 3lzc

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Biosynthetic protein
|
PDB id
|
|

|

|
3lzc
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Biosynthetic protein
|
 |
|
Title:
|
 |
Crystal structure of dph2 from pyrococcus horikoshii
|
|
Structure:
|
 |
Dph2. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Pyrococcus horikoshii. Organism_taxid: 53953. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.26Å
|
R-factor:
|
0.204
|
R-free:
|
0.240
|
|
|
Authors:
|
 |
Y.Zhang,X.Zhu,A.T.Torelli,M.Lee,B.Dzikovski,R.M.Koralewski,E.Wang, J.Freed,C.Krebs,H.Lin,S.E.Ealick
|
|
Key ref:
|
 |
Y.Zhang
et al.
(2010).
Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature,
465,
891-896.
PubMed id:
|
 |
|
Date:
|
 |
|
01-Mar-10
|
Release date:
|
23-Jun-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O58832
(DPH2_PYRHO) -
2-(3-amino-3-carboxypropyl)histidine synthase from Pyrococcus horikoshii (strain ATCC 700860 / DSM 12428 / JCM 9974 / NBRC 100139 / OT-3)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
342 a.a.
342 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.5.1.108
- 2-(3-amino-3-carboxypropyl)histidine synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-histidyl-[translation elongation factor 2] + S-adenosyl-L-methionine = 2-[(3S)-amino-3-carboxypropyl]-L-histidyl-[translation elongation factor 2] + S-methyl-5'-thioadenosine + H+
|
 |
 |
 |
 |
 |
L-histidyl-[translation elongation factor 2]
|
+
|
 S-adenosyl-L-methionine
S-adenosyl-L-methionine
|
=
|
2-[(3S)-amino-3-carboxypropyl]-L-histidyl-[translation elongation factor 2]
|
+
|
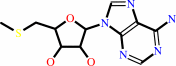 S-methyl-5'-thioadenosine
S-methyl-5'-thioadenosine
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Iron-sulfur
|
 |
 |
 |
 |
 |
 Iron-sulfur
Iron-sulfur
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nature
465:891-896
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
|
|
Y.Zhang,
X.Zhu,
A.T.Torelli,
M.Lee,
B.Dzikovski,
R.M.Koralewski,
E.Wang,
J.Freed,
C.Krebs,
S.E.Ealick,
H.Lin.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Archaeal and eukaryotic translation elongation factor 2 contain a unique
post-translationally modified histidine residue called diphthamide, which is the
target of diphtheria toxin. The biosynthesis of diphthamide was proposed to
involve three steps, with the first being the formation of a C-C bond between
the histidine residue and the 3-amino-3-carboxypropyl group of
S-adenosyl-l-methionine (SAM). However, further details of the biosynthesis
remain unknown. Here we present structural and biochemical evidence showing that
the first step of diphthamide biosynthesis in the archaeon Pyrococcus horikoshii
uses a novel iron-sulphur-cluster enzyme, Dph2. Dph2 is a homodimer and each of
its monomers can bind a [4Fe-4S] cluster. Biochemical data suggest that unlike
the enzymes in the radical SAM superfamily, Dph2 does not form the canonical
5'-deoxyadenosyl radical. Instead, it breaks the C(gamma,Met)-S bond of SAM and
generates a 3-amino-3-carboxypropyl radical. Our results suggest that P.
horikoshii Dph2 represents a previously unknown, SAM-dependent,
[4Fe-4S]-containing enzyme that catalyses unprecedented chemistry.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
F.Yan,
and
D.G.Fujimori
(2011).
RNA methylation by Radical SAM enzymes RlmN and Cfr proceeds via methylene transfer and hydride shift.
|
| |
Proc Natl Acad Sci U S A,
108,
3930-3934.
|
 |
|
|
|
|
 |
P.L.Roach
(2011).
Radicals from S-adenosylmethionine and their application to biosynthesis.
|
| |
Curr Opin Chem Biol,
15,
267-275.
|
 |
|
|
|
|
 |
S.S.Kamat,
H.J.Williams,
and
F.M.Raushel
(2011).
Intermediates in the transformation of phosphonates to phosphate by bacteria.
|
| |
Nature,
480,
570-573.
|
 |
|
|
|
|
 |
X.Zhu,
B.Dzikovski,
X.Su,
A.T.Torelli,
Y.Zhang,
S.E.Ealick,
J.H.Freed,
and
H.Lin
(2011).
Mechanistic understanding of Pyrococcus horikoshii Dph2, a [4Fe-4S] enzyme required for diphthamide biosynthesis.
|
| |
Mol Biosyst,
7,
74-81.
|
 |
|
|
|
|
 |
I.K.Blaby,
G.Phillips,
C.E.Blaby-Haas,
K.S.Gulig,
B.El Yacoubi,
and
V.de Crécy-Lagard
(2010).
Towards a systems approach in the genetic analysis of archaea: Accelerating mutant construction and phenotypic analysis in Haloferax volcanii.
|
| |
Archaea,
2010,
426239.
|
 |
|
|
|
|
 |
J.B.Broderick
(2010).
Biochemistry: A radically different enzyme.
|
| |
Nature,
465,
877-878.
|
 |
|
|
|
|
 |
M.Bucci,
C.Goodman,
and
T.L.Sheppard
(2010).
A decade of chemical biology.
|
| |
Nat Chem Biol,
6,
847-854.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

