 |
PDBsum entry 2ywc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Crystal structure of gmp synthetase from thermus thermophilus in complex with xmp
|
|
Structure:
|
 |
Gmp synthase [glutamine-hydrolyzing]. Chain: a, b, c, d. Synonym: glutamine amidotransferase, gmp synthetase. Engineered: yes
|
|
Source:
|
 |
Thermus thermophilus. Organism_taxid: 300852. Strain: hb8. Gene: guaa. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.236
|
R-free:
|
0.278
|
|
|
Authors:
|
 |
S.Baba,M.Kanagawa,S.Kuramitsu,S.Yokoyama,G.Sampei,G.Kawai,Riken Structural Genomics/proteomics Initiative (Rsgi)
|
|
Key ref:
|
 |
S.Baba
et al.
Crystal structure of gmp synthetase from thermus thermophilus.
To be published,
.
|
 |
|
Date:
|
 |
|
20-Apr-07
|
Release date:
|
23-Oct-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q5SI28
(GUAA_THET8) -
GMP synthase [glutamine-hydrolyzing] from Thermus thermophilus (strain ATCC 27634 / DSM 579 / HB8)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
503 a.a.
475 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.5.2
- Gmp synthase (glutamine-hydrolyzing).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP and GMP Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
XMP + L-glutamine + ATP + H2O = GMP + L-glutamate + AMP + diphosphate + 2 H+
|
 |
 |
 |
 |
 |
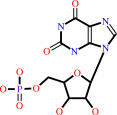 XMP
XMP
|
+
|
 L-glutamine
L-glutamine
|
+
|
 ATP
ATP
|
+
|
H2O
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
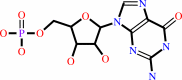 GMP
GMP
|
+
|
 L-glutamate
L-glutamate
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

