 |
PDBsum entry 2vtb

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase/DNA
|
 |
|
Title:
|
 |
Structure of cryptochrome 3 - DNA complex
|
|
Structure:
|
 |
Cryptochrome dash. Chain: a, c, d, e, f. Fragment: cryptochrome dash, residues 44-569. Synonym: cryptochrome 3. Engineered: yes. Other_details: mature protein without plastid import sequence. Cryptochrome dash. Chain: b. Fragment: cryptochrome dash, residues 44-482,484-489,490-569.
|
|
Source:
|
 |
Arabidopsis thaliana. Mouse-ear cress. Organism_taxid: 3702. Expressed in: escherichia coli. Expression_system_taxid: 562. Synthetic: yes. Homo sapiens. Human. Organism_taxid: 9606
|
|
Resolution:
|
 |
|
2.01Å
|
R-factor:
|
0.185
|
R-free:
|
0.222
|
|
|
Authors:
|
 |
R.Pokorny,T.Klar,U.Hennecke,T.Carell,A.Batschauer,L.-O.Essen
|
Key ref:

|
 |
R.Pokorny
et al.
(2008).
Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome.
Proc Natl Acad Sci U S A,
105,
21023-21027.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
13-May-08
|
Release date:
|
02-Jun-09
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q84KJ5
(CRYD_ARATH) -
Cryptochrome DASH, chloroplastic/mitochondrial from Arabidopsis thaliana
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
569 a.a.
500 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|

|

|
|
|
|
T-T-TCP-T-T
5 bases
|
|
|
|
T-T-TCP-T-T
5 bases
|
|
|
|
T-T-TCP-T-T
5 bases
|
|
|
|
T-T-TCP-T-T
5 bases
|
|
|
|
T-TCP-T-T
4 bases
|
|
|
|
T-TCP-T-T
4 bases
|
|

|
 |
 |
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.99.3
- deoxyribodipyrimidine photo-lyase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
EC 4.1.99.3
|
 |
 |
 |
 |
 |
Reaction:
|
 |
cyclobutadipyrimidine (in DNA) = 2 pyrimidine residues (in DNA)
|
 |
 |
 |
 |
 |
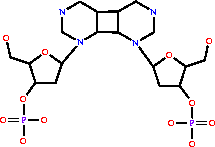 cyclobutadipyrimidine (in DNA)
cyclobutadipyrimidine (in DNA)
|
=
|
2
×
pyrimidine residues (in DNA)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
5,10-methenyltetrahydrofolate; FAD
|
 |
 |
 |
 |
 |
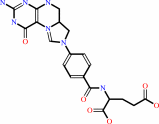 5,10-methenyltetrahydrofolate
5,10-methenyltetrahydrofolate
|
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
105:21023-21027
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome.
|
|
R.Pokorny,
T.Klar,
U.Hennecke,
T.Carell,
A.Batschauer,
L.O.Essen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
DNA photolyases and cryptochromes (cry) form a family of flavoproteins that use
light energy in the blue/UV-A region for the repair of UV-induced DNA lesions or
for signaling, respectively. Very recently, it was shown that members of the
DASH cryptochrome subclade repair specifically cyclobutane pyrimidine dimers
(CPDs) in UV-damaged single-stranded DNA. Here, we report the crystal structure
of Arabidopsis cryptochrome 3 with an in-situ-repaired CPD substrate in
single-stranded DNA. The structure shows a binding mode similar to that of
conventional DNA photolyases. Furthermore, CPD lesions in double-stranded DNA
are bound and repaired with similar efficiency as in single-stranded DNA if the
CPD lesion is present in a loop structure. Together, these data reveal that DASH
cryptochromes catalyze light-driven DNA repair like conventional photolyases but
lack an efficient flipping mechanism for interaction with CPD lesions within
duplex DNA.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Isoelectric surface potential of A. t. cry3 bound to the
single-stranded pentameric DNA containing a CPD analog. (A) Top
and side (Inset) views. (B and C) Hydration and electrostatics
of the active site in the substrate-bound state of cry3. The
black arrows in C indicate the water molecules intruded into the
active site because of replacement of a tryptophan conserved in
class I CPD photolyases by Y434.
|
 |
Figure 4.
Binding of cry3 to DNA probes containing a single T<>T dimer
in the central position. (A) Sequences and structures of probes.
The T<>T dimer is positioned within the VspI recognition site
(boxed in probe 1). Probe 1 forms a perfect duplex. In probes 2
and 3, the 5′ and 3′ thymines, respectively, of the T<>T
dimer are not hydrogen bonded to the complementary strand. In
probe 3, only one hydrogen bond is formed between the 5′
thymine of the T<>T dimer and the complementary adenine (23). In
probes 4–8, the T<>T lesion is positioned in the center of
loop structures with 2–10 base pairs. Hydrogen bonds between
complementary bases are shown as dashed lines. The upper strand
(50 nt) was labeled at the 5′ position with IRDye700 (MWG
Biotech AG) (marked with asterisk). (B and C) EMSA showing cry3
binding to probes with (B) or without (C) the central T<>T
dimer. Probes shown in A and the single-stranded control (probe
9) were mixed with cry3 (+) or with the same aliquot of buffer
(−). Arrows indicate the positions of shifted bands.
Representative gels from 2 independent experiments are shown.
(D) Quantitative binding data. Mean values and standard errors
of the 2 independent experiments are shown.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Chaves,
R.Pokorny,
M.Byrdin,
N.Hoang,
T.Ritz,
K.Brettel,
L.O.Essen,
G.T.van der Horst,
A.Batschauer,
and
M.Ahmad
(2011).
The cryptochromes: blue light photoreceptors in plants and animals.
|
| |
Annu Rev Plant Biol,
62,
335-364.
|
 |
|
|
|
|
 |
W.Gärtner,
and
P.Hegemann
(2011).
Introduction to the Symposium-in Print: Blue light effects.
|
| |
Photochem Photobiol,
87,
489-490.
|
 |
|
|
|
|
 |
A.C.Froehlich,
C.H.Chen,
W.J.Belden,
C.Madeti,
T.Roenneberg,
M.Merrow,
J.J.Loros,
and
J.C.Dunlap
(2010).
Genetic and molecular characterization of a cryptochrome from the filamentous fungus Neurospora crassa.
|
| |
Eukaryot Cell,
9,
738-750.
|
 |
|
|
|
|
 |
A.Möglich,
X.Yang,
R.A.Ayers,
and
K.Moffat
(2010).
Structure and function of plant photoreceptors.
|
| |
Annu Rev Plant Biol,
61,
21-47.
|
 |
|
|
|
|
 |
L.Sauguet,
S.Klinge,
R.L.Perera,
J.D.Maman,
and
L.Pellegrini
(2010).
Shared active site architecture between the large subunit of eukaryotic primase and DNA photolyase.
|
| |
PLoS One,
5,
e10083.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Heijde,
G.Zabulon,
F.Corellou,
T.Ishikawa,
J.Brazard,
A.Usman,
F.Sanchez,
P.Plaza,
M.Martin,
A.Falciatore,
T.Todo,
F.Y.Bouget,
and
C.Bowler
(2010).
Characterization of two members of the cryptochrome/photolyase family from Ostreococcus tauri provides insights into the origin and evolution of cryptochromes.
|
| |
Plant Cell Environ,
33,
1614-1626.
|
 |
|
|
|
|
 |
P.Xu,
H.L.Zhu,
H.B.Xu,
Z.Z.Zhang,
C.Q.Zhang,
L.X.Zhang,
and
Z.Q.Ma
(2010).
Composition and phylogenetic analysis of wheat cryptochrome gene family.
|
| |
Mol Biol Rep,
37,
825-832.
|
 |
|
|
|
|
 |
V.Exner,
C.Alexandre,
G.Rosenfeldt,
P.Alfarano,
M.Nater,
A.Caflisch,
W.Gruissem,
A.Batschauer,
and
L.Hennig
(2010).
A gain-of-function mutation of Arabidopsis cryptochrome1 promotes flowering.
|
| |
Plant Physiol,
154,
1633-1645.
|
 |
|
|
|
|
 |
E.Schleicher,
R.Bittl,
and
S.Weber
(2009).
New roles of flavoproteins in molecular cell biology: blue-light active flavoproteins studied by electron paramagnetic resonance.
|
| |
FEBS J,
276,
4290-4303.
|
 |
|
|
|
|
 |
J.Moldt,
R.Pokorny,
C.Orth,
U.Linne,
Y.Geisselbrecht,
M.A.Marahiel,
L.O.Essen,
and
A.Batschauer
(2009).
Photoreduction of the folate cofactor in members of the photolyase family.
|
| |
J Biol Chem,
284,
21670-21683.
|
 |
|
|
|
|
 |
M.Müller,
and
T.Carell
(2009).
Structural biology of DNA photolyases and cryptochromes.
|
| |
Curr Opin Struct Biol,
19,
277-285.
|
 |
|
|
|
|
 |
S.Coesel,
M.Mangogna,
T.Ishikawa,
M.Heijde,
A.Rogato,
G.Finazzi,
T.Todo,
C.Bowler,
and
A.Falciatore
(2009).
Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity.
|
| |
EMBO Rep,
10,
655-661.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

