 |
PDBsum entry 2vps

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.1.9
- trypanothione synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
spermidine + glutathione + ATP = glutathionylspermidine + ADP + phosphate + H+
|
|
2.
|
glutathionylspermidine + glutathione + ATP = trypanothione + ADP + phosphate + H+
|
|
 |
 |
 |
 |
 |
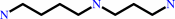 spermidine
spermidine
|
+
|
 glutathione
glutathione
|
+
|
 ATP
ATP
|
=
|
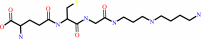 glutathionylspermidine
glutathionylspermidine
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 glutathionylspermidine
glutathionylspermidine
|
+
|
 glutathione
glutathione
|
+
|
 ATP
ATP
|
=
|
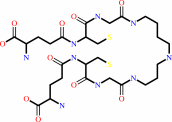 trypanothione
trypanothione
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mg(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
283:17672-17680
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Leishmania trypanothione synthetase-amidase structure reveals a basis for regulation of conflicting synthetic and hydrolytic activities.
|
|
P.K.Fyfe,
S.L.Oza,
A.H.Fairlamb,
W.N.Hunter.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The bifunctional trypanothione synthetase-amidase catalyzes biosynthesis and
hydrolysis of the glutathione-spermidine adduct trypanothione, the principal
intracellular thiol-redox metabolite in parasitic trypanosomatids. These
parasites are unique with regard to their reliance on trypanothione to determine
intracellular thiol-redox balance in defense against oxidative and chemical
stress and to regulate polyamine levels. Enzymes involved in trypanothione
biosynthesis provide essential biological activities, and those absent from
humans or for which orthologues are sufficiently distinct are attractive targets
to underpin anti-parasitic drug discovery. The structure of Leishmania major
trypanothione synthetase-amidase, determined in three crystal forms, reveals two
catalytic domains. The N-terminal domain, a cysteine, histidine-dependent
amidohydrolase/peptidase amidase, is a papain-like cysteine protease, and the
C-terminal synthetase domain displays an ATP-grasp family fold common to C:N
ligases. Modeling of substrates into each active site provides insight into the
specificity and reactivity of this unusual enzyme, which is able to catalyze
four reactions. The domain orientation is distinct from that observed in a
related bacterial glutathionylspermidine synthetase. In trypanothione
synthetase-amidase, the interactions formed by the C terminus, binding in and
restricting access to the amidase active site, suggest that the balance of
ligation and hydrolytic activity is directly influenced by the alignment of the
domains with respect to each other and implicate conformational changes with
amidase activity. The potential inhibitory role of the C terminus provides a
mechanism to control relative levels of the critical metabolites, trypanothione,
glutathionylspermidine, and spermidine in Leishmania.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIGURE 1. Ligation and hydrolytic reactions catalyzed by
trypanothione synthetase-amidase. Trypanothione is produced by
stepwise ligation of glutathione with spermidine (reaction I),
then glutathionylspermidine (reaction II). Hydrolysis of
trypanothione (reaction III) then glutathionylspermidine
(reaction IV) is performed by the N-terminal amidase domain.
|
 |
Figure 2.
FIGURE 2. Secondary, tertiary, and domain structure of
LmTSA. a, the fold. Red and black stars mark amidase and
synthetase active sites, respectively. Selected elements of
secondary structure are labeled.  1 is blue, and the
β-barrel is red. b, the subdomain structure of the ATP-grasp
synthetase domain viewed orthogonal to a. Subdomain A is colored
orange, subdomain B is blue, and subdomain C is purple. A model
of ADP (black sticks, based on structural comparisons) is
included. 1 is blue, and the
β-barrel is red. b, the subdomain structure of the ATP-grasp
synthetase domain viewed orthogonal to a. Subdomain A is colored
orange, subdomain B is blue, and subdomain C is purple. A model
of ADP (black sticks, based on structural comparisons) is
included.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2008,
283,
17672-17680)
copyright 2008.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.H.Pai,
H.J.Wu,
C.H.Lin,
and
A.H.Wang
(2011).
Structure and mechanism of Escherichia coli glutathionylspermidine amidase belonging to the family of cysteine; histidine-dependent amidohydrolases/peptidases.
|
| |
Protein Sci,
20,
557-566.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Colotti,
and
A.Ilari
(2011).
Polyamine metabolism in Leishmania: from arginine to trypanothione.
|
| |
Amino Acids,
40,
269-285.
|
 |
|
|
|
|
 |
P.K.Fyfe,
M.S.Alphey,
and
W.N.Hunter
(2010).
Structure of Trypanosoma brucei glutathione synthetase: domain and loop alterations in the catalytic cycle of a highly conserved enzyme.
|
| |
Mol Biochem Parasitol,
170,
93-99.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.S.Torrie,
S.Wyllie,
D.Spinks,
S.L.Oza,
S.Thompson,
J.R.Harrison,
I.H.Gilbert,
P.G.Wyatt,
A.H.Fairlamb,
and
J.A.Frearson
(2009).
Chemical validation of trypanothione synthetase: a potential drug target for human trypanosomiasis.
|
| |
J Biol Chem,
284,
36137-36145.
|
 |
|
|
|
|
 |
Q.Xu,
S.Sudek,
D.McMullan,
M.D.Miller,
B.Geierstanger,
D.H.Jones,
S.S.Krishna,
G.Spraggon,
B.Bursalay,
P.Abdubek,
C.Acosta,
E.Ambing,
T.Astakhova,
H.L.Axelrod,
D.Carlton,
J.Caruthers,
H.J.Chiu,
T.Clayton,
M.C.Deller,
L.Duan,
Y.Elias,
M.A.Elsliger,
J.Feuerhelm,
S.K.Grzechnik,
J.Hale,
G.W.Han,
J.Haugen,
L.Jaroszewski,
K.K.Jin,
H.E.Klock,
M.W.Knuth,
P.Kozbial,
A.Kumar,
D.Marciano,
A.T.Morse,
E.Nigoghossian,
L.Okach,
S.Oommachen,
J.Paulsen,
R.Reyes,
C.L.Rife,
C.V.Trout,
H.van den Bedem,
D.Weekes,
A.White,
G.Wolf,
C.Zubieta,
K.O.Hodgson,
J.Wooley,
A.M.Deacon,
A.Godzik,
S.A.Lesley,
and
I.A.Wilson
(2009).
Structural basis of murein peptide specificity of a gamma-D-glutamyl-l-diamino acid endopeptidase.
|
| |
Structure,
17,
303-313.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Wyllie,
S.L.Oza,
S.Patterson,
D.Spinks,
S.Thompson,
and
A.H.Fairlamb
(2009).
Dissecting the essentiality of the bifunctional trypanothione synthetase-amidase in Trypanosoma brucei using chemical and genetic methods.
|
| |
Mol Microbiol,
74,
529-540.
|
 |
|
|
|
|
 |
Y.Xiao,
D.E.McCloskey,
and
M.A.Phillips
(2009).
RNA interference-mediated silencing of ornithine decarboxylase and spermidine synthase genes in Trypanosoma brucei provides insight into regulation of polyamine biosynthesis.
|
| |
Eukaryot Cell,
8,
747-755.
|
 |
|
|
|
|
 |
E.K.Willert,
and
M.A.Phillips
(2008).
Regulated expression of an essential allosteric activator of polyamine biosynthesis in African trypanosomes.
|
| |
PLoS Pathog,
4,
e1000183.
|
 |
|
|
|
|
 |
F.Irigoín,
L.Cibils,
M.A.Comini,
S.R.Wilkinson,
L.Flohé,
and
R.Radi
(2008).
Insights into the redox biology of Trypanosoma cruzi: Trypanothione metabolism and oxidant detoxification.
|
| |
Free Radic Biol Med,
45,
733-742.
|
 |
|
|
|
|
 |
S.L.Oza,
S.Chen,
S.Wyllie,
J.K.Coward,
and
A.H.Fairlamb
(2008).
ATP-dependent ligases in trypanothione biosynthesis--kinetics of catalysis and inhibition by phosphinic acid pseudopeptides.
|
| |
FEBS J,
275,
5408-5421.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

