 |
PDBsum entry 2phd

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2phd
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure determination of a salicylate 1,2-dioxygenase from pseudaminobacter salicylatoxidans
|
|
Structure:
|
 |
Gentisate 1,2-dioxygenase. Chain: a, b, c, d. Engineered: yes
|
|
Source:
|
 |
Pseudaminobacter salicylatoxidans. Organism_taxid: 93369. Strain: bn12. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.90Å
|
R-factor:
|
0.197
|
R-free:
|
0.262
|
|
|
Authors:
|
 |
I.Matera,M.Ferraroni,F.Briganti
|
|
Key ref:
|
 |
M.Ferraroni
et al.
Crystal structure determination of a salicylate 1,2-Dioxygenase from pseudaminobacter salicylatoxidans.
To be published,
.
|
 |
|
Date:
|
 |
|
11-Apr-07
|
Release date:
|
22-Apr-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q67FT0
(Q67FT0_PSESE) -
Gentisate 1,2-dioxygenase from Pseudaminobacter salicylatoxidans
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
368 a.a.
349 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.13.11.4
- gentisate 1,2-dioxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2,5-dihydroxybenzoate + O2 = 3-maleylpyruvate + H+
|
 |
 |
 |
 |
 |
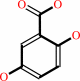 2,5-dihydroxybenzoate
2,5-dihydroxybenzoate
|
+
|
 O2
O2
|
=
|
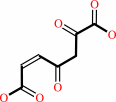 3-maleylpyruvate
3-maleylpyruvate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

