 |
PDBsum entry 2pgd

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase (choh(d)-NADP+(a))
|
PDB id
|
|

|

|
2pgd
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.44
- phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pentose Phosphate Pathway (early stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
6-phospho-D-gluconate + NADP+ = D-ribulose 5-phosphate + CO2 + NADPH
|
 |
 |
 |
 |
 |
 6-phospho-D-gluconate
6-phospho-D-gluconate
|
+
|
 NADP(+)
NADP(+)
|
=
|
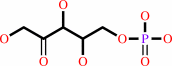 D-ribulose 5-phosphate
D-ribulose 5-phosphate
|
+
|
 CO2
CO2
|
+
|
 NADPH
NADPH
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Acta Crystallogr B
47:817-820
(1991)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of 6-phosphogluconate dehydrogenase refined at 2.5 A resolution.
|
|
M.J.Adams,
S.Gover,
R.Leaback,
C.Phillips,
D.O.Somers.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The three-dimensional structure of ovine 6-phosphogluconate dehydrogenase,
refined at 2.5 A resolution with a residual for all data of 18.5%, is reported.
This model, based on improved diffraction data and a corrected sequence,
supersedes that reported earlier. Each subunit of the dimer has three domains: a
beta-alpha-beta domain binds NADP; an all alpha domain provides much of the
dimer interface; the C-terminal tail burrows into the second subunit.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.E.Tronrud,
and
B.W.Matthews
(2009).
Sorting the chaff from the wheat at the PDB.
|
| |
Protein Sci,
18,
2-5.
|
 |
|
|
|
|
 |
M.J.Sippl
(2009).
Fold space unlimited.
|
| |
Curr Opin Struct Biol,
19,
312-320.
|
 |
|
|
|
|
 |
W.He,
Y.Wang,
W.Liu,
and
C.Z.Zhou
(2007).
Crystal structure of Saccharomyces cerevisiae 6-phosphogluconate dehydrogenase Gnd1.
|
| |
BMC Struct Biol,
7,
38.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Ceyhan,
A.Danişan,
I.H.Oğüş,
and
N.Ozer
(2005).
Purification and kinetic properties of 6-phosphogluconate dehydrogenase from rat small intestine.
|
| |
Protein J,
24,
293-301.
|
 |
|
|
|
|
 |
G.N.Goulielmos,
E.Eliopoulos,
M.Loukas,
and
S.Tsakas
(2004).
Functional constraints of 6-phosphogluconate dehydrogenase (6-PGD) based on sequence and structural information.
|
| |
J Mol Evol,
59,
358-371.
|
 |
|
|
|
|
 |
E.Warkentin,
B.Mamat,
M.Sordel-Klippert,
M.Wicke,
R.K.Thauer,
M.Iwata,
S.Iwata,
U.Ermler,
and
S.Shima
(2001).
Structures of F420H2:NADP+ oxidoreductase with and without its substrates bound.
|
| |
EMBO J,
20,
6561-6569.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.J.Adams,
G.H.Ellis,
S.Gover,
C.E.Naylor,
and
C.Phillips
(1994).
Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism.
|
| |
Structure,
2,
651-668.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Rowland,
A.K.Basak,
S.Gover,
H.R.Levy,
and
M.J.Adams
(1994).
The three-dimensional structure of glucose 6-phosphate dehydrogenase from Leuconostoc mesenteroides refined at 2.0 A resolution.
|
| |
Structure,
2,
1073-1087.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.B.Swindells
(1993).
Classification of doubly wound nucleotide binding topologies using automated loop searches.
|
| |
Protein Sci,
2,
2146-2153.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

