 |
PDBsum entry 1pgp

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase (choh(d)-NADP+(a))
|
PDB id
|
|

|

|
1pgp
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.44
- phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pentose Phosphate Pathway (early stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
6-phospho-D-gluconate + NADP+ = D-ribulose 5-phosphate + CO2 + NADPH
|
 |
 |
 |
 |
 |
6-phospho-D-gluconate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 NADP(+)
NADP(+)
|
=
|
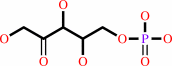 D-ribulose 5-phosphate
D-ribulose 5-phosphate
|
+
|
 CO2
CO2
|
+
|
 NADPH
NADPH
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
2:651-668
(1994)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism.
|
|
M.J.Adams,
G.H.Ellis,
S.Gover,
C.E.Naylor,
C.Phillips.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: The nicotinamide adenine dinucleotide phosphate (NADP)-dependent
oxidative decarboxylase, 6-phosphogluconate dehydrogenase, is a major source of
reduced coenzyme for synthesis. Enzymes later in the pentose phosphate pathway
convert the reaction product, ribulose 5-phosphate, to ribose 5-phosphate.
Crystallographic study of complexes with coenzyme and substrate explain the NADP
dependence which determines the enzyme's metabolic role and support the proposed
general base-general acid mechanism. RESULTS: The refined structures of binary
coenzyme/analogue complexes show that Arg33 is ordered by binding the
2'-phosphate, and provides one face of the adenine site. The nicotinamide, while
less tightly bound, is more extended when reduced than when oxidized. All
substrate binding residues are conserved; the 3-hydroxyl of 6-phosphogluconate
is hydrogen bonded to N zeta of Lys183 and the 3-hydrogen points towards the
oxidized nicotinamide. The 6-phosphate replaces a tightly bound sulphate in the
apo-enzyme. CONCLUSIONS: NADP specificity is achieved primarily by Arg33 which
binds the 2'-phosphate but, in its absence, obscures the adenine pocket. The
bound oxidized nicotinamide is syn; hydride transfer from bound substrate to the
nicotinamide si- face is achieved with a small movement of the nicotinamide
nucleotide. Lys183 may act as general base. A water bound to Gly130 in the
coenzyme domain is the most likely acid required in decarboxylation. The
dihydronicotinamide ring of NADPH competes for ligands with the 1-carboxyl of
6-phosphogluconate.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 8.
Figure 8. Stereo pair showing Arg33 in its conformation in the
apo-enzyme (blue) and in binary complexes (red), and
demonstrating its role in forming the adenine pocket. Figure
8. Stereo pair showing Arg33 in its conformation in the
apo-enzyme (blue) and in binary complexes (red), and
demonstrating its role in forming the adenine pocket.
|
 |
Figure 14.
Figure 14. Steps of the general base–acid mechanism for 6PG
oxidative decarboxylation by 6PGDH (after Berdis and Cook [21]).
Figure 14. Steps of the general base–acid mechanism for 6PG
oxidative decarboxylation by 6PGDH (after Berdis and Cook
[[3]21]).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Structure
(1994,
2,
651-668)
copyright 1994.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.F.Ruda,
G.Campbell,
V.P.Alibu,
M.P.Barrett,
R.Brenk,
and
I.H.Gilbert
(2010).
Virtual fragment screening for novel inhibitors of 6-phosphogluconate dehydrogenase.
|
| |
Bioorg Med Chem,
18,
5056-5062.
|
 |
|
|
|
|
 |
G.P.Laliotis,
I.Bizelis,
and
E.Rogdakis
(2010).
Comparative Approach of the de novo Fatty Acid Synthesis (Lipogenesis) between Ruminant and Non Ruminant Mammalian Species: From Biochemical Level to the Main Regulatory Lipogenic Genes.
|
| |
Curr Genomics,
11,
168-183.
|
 |
|
|
|
|
 |
S.Ueshima,
H.Muramatsu,
T.Nakajima,
H.Yamamoto,
S.Kato,
H.Misono,
and
S.Nagata
(2010).
Identification, Cloning, and Characterization of l-Phenylserine Dehydrogenase from Pseudomonas syringae NK-15.
|
| |
Enzyme Res,
2010,
597010.
|
 |
|
|
|
|
 |
J.Osipiuk,
M.Zhou,
S.Moy,
F.Collart,
and
A.Joachimiak
(2009).
X-ray crystal structure of GarR-tartronate semialdehyde reductase from Salmonella typhimurium.
|
| |
J Struct Funct Genomics,
10,
249-253.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Saito,
M.Robert,
H.Kochi,
G.Matsuo,
Y.Kakazu,
T.Soga,
and
M.Tomita
(2009).
Metabolite Profiling Reveals YihU as a Novel Hydroxybutyrate Dehydrogenase for Alternative Succinic Semialdehyde Metabolism in Escherichia coli.
|
| |
J Biol Chem,
284,
16442-16451.
|
 |
|
|
|
|
 |
S.Cameron,
V.P.Martini,
J.Iulek,
and
W.N.Hunter
(2009).
Geobacillus stearothermophilus 6-phosphogluconate dehydrogenase complexed with 6-phosphogluconate.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
450-454.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Sundaramoorthy,
J.Iulek,
M.P.Barrett,
O.Bidet,
G.F.Ruda,
I.H.Gilbert,
and
W.N.Hunter
(2007).
Crystal structures of a bacterial 6-phosphogluconate dehydrogenase reveal aspects of specificity, mechanism and mode of inhibition by analogues of high-energy reaction intermediates.
|
| |
FEBS J,
274,
275-286.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.He,
Y.Wang,
W.Liu,
and
C.Z.Zhou
(2007).
Crystal structure of Saccharomyces cerevisiae 6-phosphogluconate dehydrogenase Gnd1.
|
| |
BMC Struct Biol,
7,
38.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Andreeva,
and
A.G.Murzin
(2006).
Evolution of protein fold in the presence of functional constraints.
|
| |
Curr Opin Struct Biol,
16,
399-408.
|
 |
|
|
|
|
 |
L.Li,
F.S.Dworkowski,
and
P.F.Cook
(2006).
Importance in catalysis of the 6-phosphate-binding site of 6-phosphogluconate in sheep liver 6-phosphogluconate dehydrogenase.
|
| |
J Biol Chem,
281,
25568-25576.
|
 |
|
|
|
|
 |
L.Li,
and
P.F.Cook
(2006).
The 2'-phosphate of NADP is responsible for proper orientation of the nicotinamide ring in the oxidative decarboxylation reaction catalyzed by sheep liver 6-phosphogluconate dehydrogenase.
|
| |
J Biol Chem,
281,
36803-36810.
|
 |
|
|
|
|
 |
Y.Kallberg,
and
B.Persson
(2006).
Prediction of coenzyme specificity in dehydrogenases/reductases. A hidden Markov model-based method and its application on complete genomes.
|
| |
FEBS J,
273,
1177-1184.
|
 |
|
|
|
|
 |
G.N.Goulielmos,
E.Eliopoulos,
M.Loukas,
and
S.Tsakas
(2004).
Functional constraints of 6-phosphogluconate dehydrogenase (6-PGD) based on sequence and structural information.
|
| |
J Mol Evol,
59,
358-371.
|
 |
|
|
|
|
 |
N.Zamboni,
E.Fischer,
D.Laudert,
S.Aymerich,
H.P.Hohmann,
and
U.Sauer
(2004).
The Bacillus subtilis yqjI gene encodes the NADP+-dependent 6-P-gluconate dehydrogenase in the pentose phosphate pathway.
|
| |
J Bacteriol,
186,
4528-4534.
|
 |
|
|
|
|
 |
E.K.Chowdhury,
Y.Akaishi,
S.Nagata,
and
H.Misono
(2003).
Cloning and overexpression of the 3-hydroxyisobutyrate dehydrogenase gene from pseudomonas putida E23.
|
| |
Biosci Biotechnol Biochem,
67,
438-441.
|
 |
|
|
|
|
 |
G.S.Rao,
D.E.Coleman,
W.E.Karsten,
P.F.Cook,
and
B.G.Harris
(2003).
Crystallographic studies on Ascaris suum NAD-malic enzyme bound to reduced cofactor and identification of an effector site.
|
| |
J Biol Chem,
278,
38051-38058.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.L.Kavanagh,
M.Klimacek,
B.Nidetzky,
and
D.K.Wilson
(2002).
Crystal structure of Pseudomonas fluorescens mannitol 2-dehydrogenase binary and ternary complexes. Specificity and catalytic mechanism.
|
| |
J Biol Chem,
277,
43433-43442.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.K.Njau,
C.A.Herndon,
and
J.W.Hawes
(2001).
New developments in our understanding of the beta-hydroxyacid dehydrogenases.
|
| |
Chem Biol Interact,
130,
785-791.
|
 |
|
|
|
|
 |
D.Liu,
W.E.Karsten,
and
P.F.Cook
(2000).
Lysine 199 is the general acid in the NAD-malic enzyme reaction.
|
| |
Biochemistry,
39,
11955-11960.
|
 |
|
|
|
|
 |
R.E.Campbell,
S.C.Mosimann,
I.van De Rijn,
M.E.Tanner,
and
N.C.Strynadka
(2000).
The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation.
|
| |
Biochemistry,
39,
7012-7023.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.W.Au,
S.Gover,
V.M.Lam,
and
M.J.Adams
(2000).
Human glucose-6-phosphate dehydrogenase: the crystal structure reveals a structural NADP(+) molecule and provides insights into enzyme deficiency.
|
| |
Structure,
8,
293-303.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.J.Barycki,
L.K.O'Brien,
J.M.Bratt,
R.Zhang,
R.Sanishvili,
A.W.Strauss,
and
L.J.Banaszak
(1999).
Biochemical characterization and crystal structure determination of human heart short chain L-3-hydroxyacyl-CoA dehydrogenase provide insights into catalytic mechanism.
|
| |
Biochemistry,
38,
5786-5798.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Xu,
G.Bhargava,
H.Wu,
G.Loeber,
and
L.Tong
(1999).
Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme: a new class of oxidative decarboxylases.
|
| |
Structure,
7,
R877-R889.
|
 |
|
|
|
|
 |
C.C.Hwang,
A.J.Berdis,
W.E.Karsten,
W.W.Cleland,
and
P.F.Cook
(1998).
Oxidative decarboxylation of 6-phosphogluconate by 6-phosphogluconate dehydrogenase proceeds by a stepwise mechanism with NADP and APADP as oxidants.
|
| |
Biochemistry,
37,
12596-12602.
|
 |
|
|
|
|
 |
E.Tetaud,
D.R.Hall,
D.G.Gourley,
G.A.Leonard,
S.Arkison,
M.P.Barrett,
and
W.N.Hunter
(1998).
Crystallization and preliminary X-ray diffraction studies of 6-phosphogluconate dehydrogenase from Lactococcus lactis.
|
| |
Acta Crystallogr D Biol Crystallogr,
54,
1422-1424.
|
 |
|
|
|
|
 |
K.L.Britton,
Y.Asano,
and
D.W.Rice
(1998).
Crystal structure and active site location of N-(1-D-carboxylethyl)-L-norvaline dehydrogenase.
|
| |
Nat Struct Biol,
5,
593-601.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.E.Karsten,
L.Chooback,
and
P.F.Cook
(1998).
Glutamate 190 is a general acid catalyst in the 6-phosphogluconate-dehydrogenase-catalyzed reaction.
|
| |
Biochemistry,
37,
15691-15697.
|
 |
|
|
|
|
 |
A.V.Efimov
(1997).
Structural trees for protein superfamilies.
|
| |
Proteins,
28,
241-260.
|
 |
|
|
|
|
 |
C.Chothia,
T.Hubbard,
S.Brenner,
H.Barns,
and
A.Murzin
(1997).
Protein folds in the all-beta and all-alpha classes.
|
| |
Annu Rev Biophys Biomol Struct,
26,
597-627.
|
 |
|
|
|
|
 |
C.E.Bell,
T.O.Yeates,
and
D.Eisenberg
(1997).
Unusual conformation of nicotinamide adenine dinucleotide (NAD) bound to diphtheria toxin: a comparison with NAD bound to the oxidoreductase enzymes.
|
| |
Protein Sci,
6,
2084-2096.
|
 |
|
|
|
|
 |
D.Christendat,
and
J.Turnbull
(1996).
Identification of active site residues of chorismate mutase-prephenate dehydrogenase from Escherichia coli.
|
| |
Biochemistry,
35,
4468-4479.
|
 |
|
|
|
|
 |
N.Tanaka,
T.Nonaka,
M.Nakanishi,
Y.Deyashiki,
A.Hara,
and
Y.Mitsui
(1996).
Crystal structure of the ternary complex of mouse lung carbonyl reductase at 1.8 A resolution: the structural origin of coenzyme specificity in the short-chain dehydrogenase/reductase family.
|
| |
Structure,
4,
33-45.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.G.Reddy,
G.Scapin,
and
J.S.Blanchard
(1996).
Interaction of pyridine nucleotide substrates with Escherichia coli dihydrodipicolinate reductase: thermodynamic and structural analysis of binary complexes.
|
| |
Biochemistry,
35,
13294-13302.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Hanau,
M.Rippa,
M.Bertelli,
F.Dallocchio,
and
M.P.Barrett
(1996).
6-Phosphogluconate dehydrogenase from Trypanosoma brucei. Kinetic analysis and inhibition by trypanocidal drugs.
|
| |
Eur J Biochem,
240,
592-599.
|
 |
|
|
|
|
 |
P.Rowland,
A.K.Basak,
S.Gover,
H.R.Levy,
and
M.J.Adams
(1994).
The three-dimensional structure of glucose 6-phosphate dehydrogenase from Leuconostoc mesenteroides refined at 2.0 A resolution.
|
| |
Structure,
2,
1073-1087.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

