 |
PDBsum entry 2oye

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2oye
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.99.1
- prostaglandin-endoperoxide synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(5Z,8Z,11Z,14Z)-eicosatetraenoate + AH2 + 2 O2 = prostaglandin H2 + A + H2O
|
 |
 |
 |
 |
 |
(5Z,8Z,11Z,14Z)-eicosatetraenoate
Bound ligand (Het Group name = )
matches with 51.16% similarity
|
+
|
AH2
|
+
|
 2
×
O2
2
×
O2
|
=
|
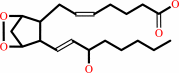 prostaglandin H2
prostaglandin H2
|
+
|
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
282:28096-28105
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis of enantioselective inhibition of cyclooxygenase-1 by S-alpha-substituted indomethacin ethanolamides.
|
|
C.A.Harman,
M.V.Turman,
K.R.Kozak,
L.J.Marnett,
W.L.Smith,
R.M.Garavito.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The modification of the nonselective nonsteroidal anti-inflammatory drug,
indomethacin, by amidation presents a promising strategy for designing novel
cyclooxygenase (COX)-2-selective inhibitors. A series of alpha-substituted
indomethacin ethanolamides, which exist as R/S-enantiomeric pairs, provides a
means to study the impact of stereochemistry on COX inhibition. Comparative
studies revealed that the R- and S-enantiomers of the alpha-substituted analogs
inhibit COX-2 with almost equal efficacy, whereas COX-1 is selectively inhibited
by the S-enantiomers. Mutagenesis studies have not been able to identify
residues that manifest the enantioselectivity in COX-1. In an effort to
understand the structural impact of chirality on COX-1 selectivity, the crystal
structures of ovine COX-1 in complexes with an enantiomeric pair of these
indomethacin ethanolamides were determined at resolutions between 2.75 and 2.85
A. These structures reveal unique, enantiomer-selective interactions within the
COX-1 side pocket region that stabilize drug binding and account for the chiral
selectivity observed with the (S)-alpha-substituted indomethacin ethanolamides.
Kinetic analysis of binding demonstrates that both inhibitors bind quickly
utilizing a two-step mechanism. However, the second binding step is readily
reversible for the R-enantiomer, whereas for the S-enantiomer, it is not. These
studies establish for the first time the structural and kinetic basis of high
affinity binding of a neutral inhibitor to COX-1 and demonstrate that the side
pocket of COX-1, previously thought to be sterically inaccessible, can serve as
a binding pocket for inhibitor association.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIGURE 1. Compound 8 bound in the COX active site of COX-1.
A, stereo view of 8 bound in COX active site with simulated
annealing omit map difference density (blue) contoured at 3  witha3Å boundary
around the ligand. Various residues within the active site are
shown with carbons for 8 shown in orange, oxygen red, nitrogen
blue, and chlorine purple. B, stereo representation of 8 bound
in the COX site in the same manner as the parent compound
indomethacin; the chlorobenzoyl group is oriented up toward the
top of the channel, the methoxy group on indole ring points
toward the side pocket (Leu-517, Phe-518, Ile-523, Gln-192, and
Ser-516), and the ethanolamide group with the R-ethyl
substitution sits next to Arg-120 and Tyr-355 at the mouth of
the active site. Carbon atoms of 8 are shown with same color
scheme as in A with the yellow dashed lines representing various
interactions (hydrophilic) between ligand and protein residues.
The hydroxyl group of the ethanolamide group makes a hydrogen
bond with Arg-120 and Glu-524, whereas the (R)-ethyl group is
positioned just outside the mouth of the active site. Refinement
statistics for both structures are shown in Table 2. All figures
presented here were created using the program PyMOL. witha3Å boundary
around the ligand. Various residues within the active site are
shown with carbons for 8 shown in orange, oxygen red, nitrogen
blue, and chlorine purple. B, stereo representation of 8 bound
in the COX site in the same manner as the parent compound
indomethacin; the chlorobenzoyl group is oriented up toward the
top of the channel, the methoxy group on indole ring points
toward the side pocket (Leu-517, Phe-518, Ile-523, Gln-192, and
Ser-516), and the ethanolamide group with the R-ethyl
substitution sits next to Arg-120 and Tyr-355 at the mouth of
the active site. Carbon atoms of 8 are shown with same color
scheme as in A with the yellow dashed lines representing various
interactions (hydrophilic) between ligand and protein residues.
The hydroxyl group of the ethanolamide group makes a hydrogen
bond with Arg-120 and Glu-524, whereas the (R)-ethyl group is
positioned just outside the mouth of the active site. Refinement
statistics for both structures are shown in Table 2. All figures
presented here were created using the program PyMOL.
|
 |
Figure 2.
FIGURE 2. Compound 9 bound in the COX active site of COX-1.
A, stereo view of 9 fitted into simulated annealing omit map
difference density (blue) contoured at 3  witha3Å boundary
around the ligand. Carbon atoms of 9 are colored green with
heteroatoms colored the same as in 1. B, chlorobenzoyl group of
9 is oriented toward the mouth of the active site; the methoxy
group of indole ring is oriented up into the top of the channel,
and the ethanolamide group with the (S)-ethyl substitution lies
within the side pocket region of the channel. Yellow dashed
lines represent various interactions (hydrophobic and
hydrophilic) between ligand and protein. The hydroxyl group of
the ethanolamide is oriented near the hydrophilic region (cyan)
of the side pocket making hydrogen bonds with residues Gln-192
and His-90 whereas the S-ethyl group is oriented into the more
hydrophobic region (pink) of the side pocket making van der
Waals interactions with Ile-523, Phe-518, and Ile-517. For
clarity, an additional polar interaction between Arg-120 and the
carbonyl of the chlorobenzoyl moiety is not shown. witha3Å boundary
around the ligand. Carbon atoms of 9 are colored green with
heteroatoms colored the same as in 1. B, chlorobenzoyl group of
9 is oriented toward the mouth of the active site; the methoxy
group of indole ring is oriented up into the top of the channel,
and the ethanolamide group with the (S)-ethyl substitution lies
within the side pocket region of the channel. Yellow dashed
lines represent various interactions (hydrophobic and
hydrophilic) between ligand and protein. The hydroxyl group of
the ethanolamide is oriented near the hydrophilic region (cyan)
of the side pocket making hydrogen bonds with residues Gln-192
and His-90 whereas the S-ethyl group is oriented into the more
hydrophobic region (pink) of the side pocket making van der
Waals interactions with Ile-523, Phe-518, and Ile-517. For
clarity, an additional polar interaction between Arg-120 and the
carbonyl of the chlorobenzoyl moiety is not shown.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2007,
282,
28096-28105)
copyright 2007.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.Parisini,
P.Metrangolo,
T.Pilati,
G.Resnati,
and
G.Terraneo
(2011).
Halogen bonding in halocarbon-protein complexes: a structural survey.
|
| |
Chem Soc Rev,
40,
2267-2278.
|
 |
|
|
|
|
 |
G.Rimon,
R.S.Sidhu,
D.A.Lauver,
J.Y.Lee,
N.P.Sharma,
C.Yuan,
R.A.Frieler,
R.C.Trievel,
B.R.Lucchesi,
and
W.L.Smith
(2010).
Coxibs interfere with the action of aspirin by binding tightly to one monomer of cyclooxygenase-1.
|
| |
Proc Natl Acad Sci U S A,
107,
28-33.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.S.Deeb,
C.Cheung,
T.Nuriel,
B.D.Lamon,
R.K.Upmacis,
S.S.Gross,
and
D.P.Hajjar
(2010).
Physical evidence for substrate binding in preventing cyclooxygenase inactivation under nitrative stress.
|
| |
J Am Chem Soc,
132,
3914-3922.
|
 |
|
|
|
|
 |
C.Yuan,
R.S.Sidhu,
D.V.Kuklev,
Y.Kado,
M.Wada,
I.Song,
and
W.L.Smith
(2009).
Cyclooxygenase Allosterism, Fatty Acid-mediated Cross-talk between Monomers of Cyclooxygenase Homodimers.
|
| |
J Biol Chem,
284,
10046-10055.
|
 |
|
|
|
|
 |
M.J.Walters,
A.L.Blobaum,
P.J.Kingsley,
A.S.Felts,
G.A.Sulikowski,
and
L.J.Marnett
(2009).
The influence of double bond geometry in the inhibition of cyclooxygenases by sulindac derivatives.
|
| |
Bioorg Med Chem Lett,
19,
3271-3274.
|
 |
|
|
|
|
 |
V.F.Roche
(2009).
A receptor-grounded approach to teaching nonsteroidal antiinflammatory drug chemistry and structure-activity relationships.
|
| |
Am J Pharm Educ,
73,
143.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

