
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|





 (+ 1 more)
355 a.a.
(+ 1 more)
355 a.a.
|
 |

|

|

|

|

|

|

|
 337 a.a.
337 a.a.
|
 |

|

|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Glutamate 5-kinase from escherichia coli complexed with glutamate
|
|
Structure:
|
 |
Glutamate 5-kinase. Chain: a, b, c, d, e, f, g, h. Synonym: gamma-glutamyl kinase, gk. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562. Strain: dh5alpha. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.90Å
|
R-factor:
|
0.199
|
R-free:
|
0.246
|
|
|
Authors:
|
 |
C.Marco-Marin,F.Gil-Ortiz,I.Perez-Arellano,J.Cervera,I.Fita,V.Rubio
|
Key ref:

|
 |
C.Marco-Marín
et al.
(2007).
A novel two-domain architecture within the amino acid kinase enzyme family revealed by the crystal structure of Escherichia coli glutamate 5-kinase.
J Mol Biol,
367,
1431-1446.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
19-Sep-06
|
Release date:
|
06-Mar-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F, G, H:
E.C.2.7.2.11
- glutamate 5-kinase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Proline Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-glutamate + ATP = L-glutamyl 5-phosphate + ADP
|
 |
 |
 |
 |
 |
 L-glutamate
L-glutamate
|
+
|
ATP
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
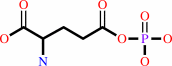 L-glutamyl 5-phosphate
L-glutamyl 5-phosphate
|
+
|
 ADP
ADP
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
367:1431-1446
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
A novel two-domain architecture within the amino acid kinase enzyme family revealed by the crystal structure of Escherichia coli glutamate 5-kinase.
|
|
C.Marco-Marín,
F.Gil-Ortiz,
I.Pérez-Arellano,
J.Cervera,
I.Fita,
V.Rubio.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Glutamate 5-kinase (G5K) makes the highly unstable product glutamyl 5-phosphate
(G5P) in the initial, controlling step of proline/ornithine synthesis, being
feedback-inhibited by proline or ornithine, and causing, when defective,
clinical hyperammonaemia. We determined two crystal structures of G5K from
Escherichia coli, at 2.9 A and 2.5 A resolution, complexed with glutamate and
sulphate, or with G5P, sulphate and the proline analogue 5-oxoproline. E. coli
G5K presents a novel tetrameric (dimer of dimers) architecture. Each subunit
contains a 257 residue AAK domain, typical of acylphosphate-forming enzymes,
with characteristic alpha(3)beta(8)alpha(4) sandwich topology. This domain is
responsible for catalysis and proline inhibition, and has a crater on the beta
sheet C-edge that hosts the active centre and bound 5-oxoproline. Each subunit
contains a 93 residue C-terminal PUA domain, typical of RNA-modifying enzymes,
which presents the characteristic beta(5)beta(4) sandwich fold and three alpha
helices. The AAK and PUA domains of one subunit associate non-canonically in the
dimer with the same domains of the other subunit, leaving a negatively charged
hole between them that hosts two Mg ions in one crystal, in line with the G5K
requirement for free Mg. The tetramer, formed by two dimers interacting
exclusively through their AAK domains, is flat and elongated, and has in each
face, pericentrically, two exposed active centres in alternate subunits. This
would permit the close apposition of two active centres of bacterial
glutamate-5-phosphate reductase (the next enzyme in the
proline/ornithine-synthesising route), supporting the postulated channelling of
G5P. The structures clarify substrate binding and catalysis, justify the high
glutamate specificity, explain the effects of known point mutations, and support
the binding of proline near glutamate. Proline binding may trigger the movement
of a loop that encircles glutamate, and which participates in a hydrogen bond
network connecting active centres, which is possibly involved in the
cooperativity for glutamate.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Pathway of proline synthesis in microorganisms and
plants, and of ornithine synthesis in mammals. Enzymes are
enclosed in grey boxes. Feed-back inhibition of microbial and
plant G5Ks by proline and of animal G5Ks by ornithine is
indicated with broken arrows. The dotted arrow indicates the
spontaneous cyclization of G5P to 5-oxoproline that is an
abortive side-reaction.
|
 |
Figure 9.
Figure 9. Possible interaction between the glutamate 5-kinase
(G5K) and the glutamyl 5-phosphate reductase (G5PR). Surface
representation of two perpendicular views of the G5K tetramer,
with the substrates in space-filling representation. A dimer of
the G5PR from T. maritima is shown (ribbon representation) with
one subunit (orange) presenting the open conformation as
observed in the crystal structure of this enzyme in the absence
of substrates (PDB 1O20), and with the other subunit (yellow)
presenting a closed conformation modelled from class 3 aldehyde
dehydrogenase complexed to NAD (PDB 1AD3). The catalytic and
NADPH-binding domains of G5PR are identified.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2007,
367,
1431-1446)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Veeravalli,
D.Boyd,
B.L.Iverson,
J.Beckwith,
and
G.Georgiou
(2011).
Laboratory evolution of glutathione biosynthesis reveals natural compensatory pathways.
|
| |
Nat Chem Biol,
7,
101-105.
|
 |
|
|
|
|
 |
I.Pérez-Arellano,
F.Carmona-Alvarez,
A.I.Martínez,
J.Rodríguez-Díaz,
and
J.Cervera
(2010).
Pyrroline-5-carboxylate synthase and proline biosynthesis: from osmotolerance to rare metabolic disease.
|
| |
Protein Sci,
19,
372-382.
|
 |
|
|
|
|
 |
I.Pérez-Arellano,
and
J.Cervera
(2010).
Glutamate kinase from Thermotoga maritima: characterization of a thermophilic enzyme for proline biosynthesis.
|
| |
Extremophiles,
14,
409-415.
|
 |
|
|
|
|
 |
M.F.Mabanglo,
H.L.Schubert,
M.Chen,
C.P.Hill,
and
C.D.Poulter
(2010).
X-ray structures of isopentenyl phosphate kinase.
|
| |
ACS Chem Biol,
5,
517-527.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Dellas,
and
J.P.Noel
(2010).
Mutation of archaeal isopentenyl phosphate kinase highlights mechanism and guides phosphorylation of additional isoprenoid monophosphates.
|
| |
ACS Chem Biol,
5,
589-601.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.D.Copley
(2009).
Evolution of efficient pathways for degradation of anthropogenic chemicals.
|
| |
Nat Chem Biol,
5,
559-566.
|
 |
|
|
|
|
 |
T.Kaino,
and
H.Takagi
(2009).
Proline as a stress protectant in the yeast Saccharomyces cerevisiae: effects of trehalose and PRO1 gene expression on stress tolerance.
|
| |
Biosci Biotechnol Biochem,
73,
2131-2135.
|
 |
|
|
|
|
 |
D.Shi,
V.Sagar,
Z.Jin,
X.Yu,
L.Caldovic,
H.Morizono,
N.M.Allewell,
and
M.Tuchman
(2008).
The crystal structure of N-acetyl-L-glutamate synthase from Neisseria gonorrhoeae provides insights into mechanisms of catalysis and regulation.
|
| |
J Biol Chem,
283,
7176-7184.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Takagi
(2008).
Proline as a stress protectant in yeast: physiological functions, metabolic regulations, and biotechnological applications.
|
| |
Appl Microbiol Biotechnol,
81,
211-223.
|
 |
|
|
|
|
 |
S.Pakhomova,
S.G.Bartlett,
A.Augustus,
T.Kuzuyama,
and
M.E.Newcomer
(2008).
Crystal Structure of Fosfomycin Resistance Kinase FomA from Streptomyces wedmorensis.
|
| |
J Biol Chem,
283,
28518-28526.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Pérez-Arellano,
J.Gallego,
and
J.Cervera
(2007).
The PUA domain - a structural and functional overview.
|
| |
FEBS J,
274,
4972-4984.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

