 |
PDBsum entry 2i4l

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.15
- proline--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Pro) + L-proline + ATP = L-prolyl-tRNA(Pro) + AMP + diphosphate
|
 |
 |
 |
 |
 |
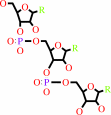 tRNA(Pro)
tRNA(Pro)
|
+
|
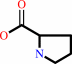 L-proline
L-proline
|
+
|
 ATP
ATP
|
=
|
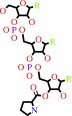 L-prolyl-tRNA(Pro)
L-prolyl-tRNA(Pro)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
14:1511-1525
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of two bacterial prolyl-tRNA synthetases with and without a cis-editing domain.
|
|
T.Crepin,
A.Yaremchuk,
M.Tukalo,
S.Cusack.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Prolyl-tRNA synthetases (ProRSs) are unique among synthetases in that they have
diverse architectures, notably the variable presence of a cis-editing domain
homologous to the freestanding deacylase proteins YbaK and ProX. Here, we
describe crystal structures of two bacterial ProRSs from the pathogen
Enterococcus faecalis, which possesses an editing domain, and from
Rhodopseudomonas palustris, which does not. We compare the overall structure and
binding mode of ATP and prolyl-adenylate with those of the
archael/eukaryote-type ProRS from Thermus thermophilus. Although structurally
more homologous to YbaK, which preferentially hydrolyzes Cys-tRNA(Pro), the
editing domain of E. faecalis ProRS possesses key elements similar to ProX, with
which it shares the activity of hydrolyzing Ala-tRNA(Pro). The structures give
insight into the complex evolution of ProRSs, the mechanism of editing, and
structural differences between prokaryotic- and eukaryotic-type ProRSs that can
be exploited for antibiotic design.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. Interactions of ProRSs with ATP
(A–C) Active
sites of (A) R. palustris, (B) E. faecalis, and (C) T.
thermophilus ProRS with bound ATP (predominantly pink molecule),
showing hydrogen bonds with key interacting residues. The
insertion domains are in magenta (in [A] and [B]), and the
eukaryote/archae-type-specific C-terminal domain is in yellow
(in [C]). Note the functionally equivalent roles of Glu218 in
PrsRp and PrsEf with the conserved carboxy terminus (Tyr477) in
PrsTt. In each case, the proline-binding loop is in the open
conformation.
|
 |
Figure 4.
Figure 4. Interactions of ProRSs with the Prolyl-Adenylate
Analog, ProAMS
(A–C) ProAMS (predominantly gray molecule)
bound in the active sites of (A) R. palustris, (B) E. faecalis,
and (C) T. thermophilus ProRS, showing key hydrogen bonds to the
proline and sulfate moieties. Class II synthetase conserved
motifs 1, 2, and 3 are shown in green, cyan, and red,
respectively, and the TXE loop is shown in gold. On the
proline-binding loop (violet), which is in the closed
conformation, hydrophobic residues Ile202, Met202, and Phe205
play equivalent roles in PrsRp, PrsEf, and PrsTt, respectively.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Structure
(2006,
14,
1511-1525)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Minajigi,
B.Deng,
and
C.S.Francklyn
(2011).
Fidelity escape by the unnatural amino acid β-hydroxynorvaline: an efficient substrate for Escherichia coli threonyl-tRNA synthetase with toxic effects on growth.
|
| |
Biochemistry,
50,
1101-1109.
|
 |
|
|
|
|
 |
A.Y.Mulkidjanian,
and
M.Y.Galperin
(2009).
On the origin of life in the Zinc world. 2. Validation of the hypothesis on the photosynthesizing zinc sulfide edifices as cradles of life on Earth.
|
| |
Biol Direct,
4,
27.
|
 |
|
|
|
|
 |
J.Ling,
B.R.So,
S.S.Yadavalli,
H.Roy,
S.Shoji,
K.Fredrick,
K.Musier-Forsyth,
and
M.Ibba
(2009).
Resampling and editing of mischarged tRNA prior to translation elongation.
|
| |
Mol Cell,
33,
654-660.
|
 |
|
|
|
|
 |
C.S.Francklyn
(2008).
DNA polymerases and aminoacyl-tRNA synthetases: shared mechanisms for ensuring the fidelity of gene expression.
|
| |
Biochemistry,
47,
11695-11703.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

