 |
PDBsum entry 2fpp

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Crystal structure of pig gtp-specific succinyl-coa synthetase from polyethylene glycol with chloride ions
|
|
Structure:
|
 |
Succinyl-coa ligase [gdp-forming] alpha-chain, mitochondrial. Chain: a. Synonym: succinyl-coa synthetase, alpha chain, scs-alpha. Engineered: yes. Succinyl-coa ligase [gdp-forming] beta-chain, mitochondrial. Chain: b. Synonym: succinyl-coa synthetase, betag chain, scs-betag, gtp-
|
|
Source:
|
 |
Sus scrofa. Pig. Organism_taxid: 9823. Gene: suclg1. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008. Gene: suclg2.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.35Å
|
R-factor:
|
0.192
|
R-free:
|
0.259
|
|
|
Authors:
|
 |
M.E.Fraser,K.Hayakawa,M.S.Hume,D.G.Ryan,E.R.Brownie
|
Key ref:

|
 |
M.E.Fraser
et al.
(2006).
Interactions of GTP with the ATP-grasp domain of GTP-specific succinyl-CoA synthetase.
J Biol Chem,
281,
11058-11065.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
16-Jan-06
|
Release date:
|
21-Feb-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains A, B:
E.C.6.2.1.4
- succinate--CoA ligase (GDP-forming).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Citric acid cycle
|
 |
 |
 |
 |
 |
Reaction:
|
 |
GTP + succinate + CoA = succinyl-CoA + GDP + phosphate
|
 |
 |
 |
 |
 |
 GTP
GTP
|
+
|
 succinate
succinate
|
+
|
 CoA
CoA
|
=
|
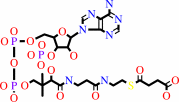 succinyl-CoA
succinyl-CoA
|
+
|
 GDP
GDP
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chain A:
E.C.6.2.1.5
- succinate--CoA ligase (ADP-forming).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
succinate + ATP + CoA = succinyl-CoA + ADP + phosphate
|
 |
 |
 |
 |
 |
 succinate
succinate
|
+
|
 ATP
ATP
|
+
|
 CoA
CoA
|
=
|
 succinyl-CoA
succinyl-CoA
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
281:11058-11065
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Interactions of GTP with the ATP-grasp domain of GTP-specific succinyl-CoA synthetase.
|
|
M.E.Fraser,
K.Hayakawa,
M.S.Hume,
D.G.Ryan,
E.R.Brownie.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Two isoforms of succinyl-CoA synthetase exist in mammals, one specific for ATP
and the other for GTP. The GTP-specific form of pig succinyl-CoA synthetase has
been crystallized in the presence of GTP and the structure determined to 2.1 A
resolution. GTP is bound in the ATP-grasp domain, where interactions of the
guanine base with a glutamine residue (Gln-20beta) and with backbone atoms
provide the specificity. The gamma-phosphate interacts with the side chain of an
arginine residue (Arg-54beta) and with backbone amide nitrogen atoms, leading to
tight interactions between the gamma-phosphate and the protein. This contrasts
with the structures of ATP bound to other members of the family of ATP-grasp
proteins where the gamma-phosphate is exposed, free to react with the other
substrate. To test if GDP would interact with GTP-specific succinyl-CoA
synthetase in the same way that ADP interacts with other members of the family
of ATP-grasp proteins, the structure of GDP bound to GTP-specific succinyl-CoA
synthetase was also determined. A comparison of the conformations of GTP and GDP
shows that the bases adopt the same position but that changes in conformation of
the ribose moieties and the alpha- and beta-phosphates allow the gamma-phosphate
to interact with the arginine residue and amide nitrogen atoms in GTP, while the
beta-phosphate interacts with these residues in GDP. The complex of GTP with
succinyl-CoA synthetase shows that the enzyme is able to protect GTP from
hydrolysis when the active-site histidine residue is not in position to be
phosphorylated.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIGURE 1. A, ribbon diagram of pig GTP-specific SCS showing
the location of the GTP-binding site. The  -subunit is green, the -subunit is green, the
 -subunit is yellow,
except for the T-loop, which is highlighted in magenta. GTP, the
potassium ion, and the side chain of the phosphorylated
histidine residue, His-259 -subunit is yellow,
except for the T-loop, which is highlighted in magenta. GTP, the
potassium ion, and the side chain of the phosphorylated
histidine residue, His-259  , are drawn as stick
models and colored according to atom type: red for oxygen,
yellow for carbon, blue for nitrogen, green for phosphorus, and
turquoise for potassium. B, stereo view of the electron density
for GTP and the potassium ion, including nearby residues of the
ATP-grasp domain of pig GTP-specific SCS. The F[o] - F[c], , are drawn as stick
models and colored according to atom type: red for oxygen,
yellow for carbon, blue for nitrogen, green for phosphorus, and
turquoise for potassium. B, stereo view of the electron density
for GTP and the potassium ion, including nearby residues of the
ATP-grasp domain of pig GTP-specific SCS. The F[o] - F[c],  [c]
electron density map calculated without GTP and the potassium
ion is contoured at 3 [c]
electron density map calculated without GTP and the potassium
ion is contoured at 3  . C, stereo view of the
electron density for GDP and the potassium ion, including nearby
residues of the ATP-grasp domain of pig GTP-specific SCS. The
F[o] - F[c], . C, stereo view of the
electron density for GDP and the potassium ion, including nearby
residues of the ATP-grasp domain of pig GTP-specific SCS. The
F[o] - F[c],  [c] electron density
map calculated without GDP and the potassium ion is contoured at
3 [c] electron density
map calculated without GDP and the potassium ion is contoured at
3  .
The same atom colors were used in B and C as described for A.
All parts of the figure were drawn using the program RASTER3D
(58), and B and C also used the program XFIT (47). .
The same atom colors were used in B and C as described for A.
All parts of the figure were drawn using the program RASTER3D
(58), and B and C also used the program XFIT (47).
|
 |
Figure 3.
FIGURE 3. Stereo views of the superpositions of GTP bound
to pig GTP-specific SCS (black) with ADP bound to E. coli SCS
(gray) (Protein Data Bank (43) identifier 1cqi (7)) (A), GDP
bound to pig GTP-specific SCS (gray) (B), and ATP bound to
glycinamide ribonucleotide transformylase (gray) (PDB identifier
1kj8) (54) (C). The superpositions were based on structurally
similar residues and performed using the program O (51) with a
cutoff of 3.8 Å. Possible hydrogen-bonding interactions
and ionic interactions between GTP and the pig GTP-specific SCS
are represented by black dashed lines.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2006,
281,
11058-11065)
copyright 2006.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.Singh,
J.Lemire,
R.J.Mailloux,
D.Chénier,
R.Hamel,
and
V.D.Appanna
(2009).
An ATP and oxalate generating variant tricarboxylic acid cycle counters aluminum toxicity in pseudomonas fluorescens.
|
| |
PLoS One,
4,
e7344.
|
 |
|
|
|
|
 |
K.Hamblin,
D.M.Standley,
M.B.Rogers,
A.Stechmann,
A.J.Roger,
R.Maytum,
and
M.van der Giezen
(2008).
Localization and nucleotide specificity of Blastocystis succinyl-CoA synthetase.
|
| |
Mol Microbiol,
68,
1395-1405.
|
 |
|
|
|
|
 |
E.Hidber,
E.R.Brownie,
K.Hayakawa,
and
M.E.Fraser
(2007).
Participation of Cys123alpha of Escherichia coli succinyl-CoA synthetase in catalysis.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
876-884.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.A.Joyce,
E.R.Brownie,
K.Hayakawa,
and
M.E.Fraser
(2007).
Cloning, expression, purification, crystallization and preliminary X-ray analysis of Thermus aquaticus succinyl-CoA synthetase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
399-402.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |
|

