 |
PDBsum entry 2fmt

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Complex (methyltransferase/tRNA)
|
PDB id
|
|

|

|
2fmt
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.2.9
- methionyl-tRNA formyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Folate Coenzymes
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-methionyl-tRNA(fMet) + (6R)-10-formyltetrahydrofolate = N-formyl-L- methionyl-tRNA(fMet) + (6S)-5,6,7,8-tetrahydrofolate + H+
|
 |
 |
 |
 |
 |
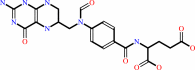 10-formyltetrahydrofolate
10-formyltetrahydrofolate
|
+
|
L-methionyl-tRNA(fMet)
|
=
|
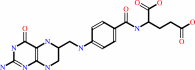 tetrahydrofolate
tetrahydrofolate
|
+
|
N-formylmethionyl-tRNA(fMet)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Embo J
17:6819-6826
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of methionyl-tRNAfMet transformylase complexed with the initiator formyl-methionyl-tRNAfMet.
|
|
E.Schmitt,
M.Panvert,
S.Blanquet,
Y.Mechulam.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of Escherichia coli methionyl-tRNAfMet transformylase
complexed with formyl-methionyl-tRNAfMet was solved at 2.8 A resolution. The
formylation reaction catalyzed by this enzyme irreversibly commits
methionyl-tRNAfMet to initiation of translation in eubacteria. In the
three-dimensional model, the methionyl-tRNAfMet formyltransferase fills in the
inside of the L-shaped tRNA molecule on the D-stem side. The anticodon stem and
loop are away from the protein. An enzyme loop is wedged in the major groove of
the acceptor helix. As a result, the C1-A72 mismatch characteristic of the
initiator tRNA is split and the 3' arm bends inside the active centre. This
recognition mechanism is markedly distinct from that of elongation factor Tu,
which binds the acceptor arm of aminoacylated elongator tRNAs on the T-stem side.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.Schmitt,
M.Panvert,
C.Lazennec-Schurdevin,
P.D.Coureux,
J.Perez,
A.Thompson,
and
Y.Mechulam
(2012).
Structure of the ternary initiation complex aIF2-GDPNP-methionylated initiator tRNA.
|
| |
Nat Struct Mol Biol,
19,
450-454.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Tsuboi,
H.Morita,
Y.Nozaki,
K.Akama,
T.Ueda,
K.Ito,
K.H.Nierhaus,
and
N.Takeuchi
(2009).
EF-G2mt is an exclusive recycling factor in mammalian mitochondrial protein synthesis.
|
| |
Mol Cell,
35,
502-510.
|
 |
|
|
|
|
 |
S.A.Krupenko
(2009).
FDH: an aldehyde dehydrogenase fusion enzyme in folate metabolism.
|
| |
Chem Biol Interact,
178,
84-93.
|
 |
|
|
|
|
 |
A.Wang,
N.Winblade Nairn,
R.S.Johnson,
D.A.Tirrell,
and
K.Grabstein
(2008).
Processing of N-terminal unnatural amino acids in recombinant human interferon-beta in Escherichia coli.
|
| |
Chembiochem,
9,
324-330.
|
 |
|
|
|
|
 |
P.Barraud,
E.Schmitt,
Y.Mechulam,
F.Dardel,
and
C.Tisné
(2008).
A unique conformation of the anticodon stem-loop is associated with the capacity of tRNAfMet to initiate protein synthesis.
|
| |
Nucleic Acids Res,
36,
4894-4901.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Ghosh,
and
S.Vishveshwara
(2007).
A study of communication pathways in methionyl- tRNA synthetase by molecular dynamics simulations and structure network analysis.
|
| |
Proc Natl Acad Sci U S A,
104,
15711-15716.
|
 |
|
|
|
|
 |
A.Weixlbaumer,
S.Petry,
C.M.Dunham,
M.Selmer,
A.C.Kelley,
and
V.Ramakrishnan
(2007).
Crystal structure of the ribosome recycling factor bound to the ribosome.
|
| |
Nat Struct Mol Biol,
14,
733-737.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Tyagi,
and
D.H.Mathews
(2007).
Predicting helical coaxial stacking in RNA multibranch loops.
|
| |
RNA,
13,
939-951.
|
 |
|
|
|
|
 |
K.Suto,
Y.Shimizu,
K.Watanabe,
T.Ueda,
S.Fukai,
O.Nureki,
and
K.Tomita
(2006).
Crystal structures of leucyl/phenylalanyl-tRNA-protein transferase and its complex with an aminoacyl-tRNA analog.
|
| |
EMBO J,
25,
5942-5950.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Selmer,
C.M.Dunham,
F.V.Murphy,
A.Weixlbaumer,
S.Petry,
A.C.Kelley,
J.R.Weir,
and
V.Ramakrishnan
(2006).
Structure of the 70S ribosome complexed with mRNA and tRNA.
|
| |
Science,
313,
1935-1942.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.S.Laursen,
H.P.Sørensen,
K.K.Mortensen,
and
H.U.Sperling-Petersen
(2005).
Initiation of protein synthesis in bacteria.
|
| |
Microbiol Mol Biol Rev,
69,
101-123.
|
 |
|
|
|
|
 |
G.J.Williams,
S.D.Breazeale,
C.R.Raetz,
and
J.H.Naismith
(2005).
Structure and function of both domains of ArnA, a dual function decarboxylase and a formyltransferase, involved in 4-amino-4-deoxy-L-arabinose biosynthesis.
|
| |
J Biol Chem,
280,
23000-23008.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Z.Gatzeva-Topalova,
A.P.May,
and
M.C.Sousa
(2005).
Crystal structure and mechanism of the Escherichia coli ArnA (PmrI) transformylase domain. An enzyme for lipid A modification with 4-amino-4-deoxy-L-arabinose and polymyxin resistance.
|
| |
Biochemistry,
44,
5328-5338.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.D.Breazeale,
A.A.Ribeiro,
A.L.McClerren,
and
C.R.Raetz
(2005).
A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-Amino-4-deoxy-L-arabinose. Identification and function oF UDP-4-deoxy-4-formamido-L-arabinose.
|
| |
J Biol Chem,
280,
14154-14167.
|
 |
|
|
|
|
 |
A.A.Chumanevich,
S.A.Krupenko,
and
C.Davies
(2004).
The crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase: mechanism of hydrolysis and its interplay with the dehydrogenase domain.
|
| |
J Biol Chem,
279,
14355-14364.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Randau,
S.Schauer,
A.Ambrogelly,
J.C.Salazar,
J.Moser,
S.Sekine,
S.Yokoyama,
D.Söll,
and
D.Jahn
(2004).
tRNA recognition by glutamyl-tRNA reductase.
|
| |
J Biol Chem,
279,
34931-34937.
|
 |
|
|
|
|
 |
P.S.Klosterman,
D.K.Hendrix,
M.Tamura,
S.R.Holbrook,
and
S.E.Brenner
(2004).
Three-dimensional motifs from the SCOR, structural classification of RNA database: extruded strands, base triples, tetraloops and U-turns.
|
| |
Nucleic Acids Res,
32,
2342-2352.
|
 |
|
|
|
|
 |
S.Biarrotte-Sorin,
A.P.Maillard,
J.Delettré,
W.Sougakoff,
M.Arthur,
and
C.Mayer
(2004).
Crystal structures of Weissella viridescens FemX and its complex with UDP-MurNAc-pentapeptide: insights into FemABX family substrates recognition.
|
| |
Structure,
12,
257-267.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Stortchevoi,
U.Varshney,
and
U.L.RajBhandary
(2003).
Common location of determinants in initiator transfer RNAs for initiator-elongator discrimination in bacteria and in eukaryotes.
|
| |
J Biol Chem,
278,
17672-17679.
|
 |
|
|
|
|
 |
H.Yang,
F.Jossinet,
N.Leontis,
L.Chen,
J.Westbrook,
H.Berman,
and
E.Westhof
(2003).
Tools for the automatic identification and classification of RNA base pairs.
|
| |
Nucleic Acids Res,
31,
3450-3460.
|
 |
|
|
|
|
 |
A.D.Wolfson,
and
O.C.Uhlenbeck
(2002).
Modulation of tRNAAla identity by inorganic pyrophosphatase.
|
| |
Proc Natl Acad Sci U S A,
99,
5965-5970.
|
 |
|
|
|
|
 |
C.Mayer,
and
U.L.RajBhandary
(2002).
Conformational change of Escherichia coli initiator methionyl-tRNA(fMet) upon binding to methionyl-tRNA formyl transferase.
|
| |
Nucleic Acids Res,
30,
2844-2850.
|
 |
|
|
|
|
 |
J.Pei,
and
N.V.Grishin
(2002).
Breaking the singleton of germination protease.
|
| |
Protein Sci,
11,
691-697.
|
 |
|
|
|
|
 |
T.H.Tan,
N.Bochud-Allemann,
E.K.Horn,
and
A.Schneider
(2002).
Eukaryotic-type elongator tRNAMet of Trypanosoma brucei becomes formylated after import into mitochondria.
|
| |
Proc Natl Acad Sci U S A,
99,
1152-1157.
|
 |
|
|
|
|
 |
V.Ramesh,
C.Köhrer,
and
U.L.RajBhandary
(2002).
Expression of Escherichia coli methionyl-tRNA formyltransferase in Saccharomyces cerevisiae leads to formylation of the cytoplasmic initiator tRNA and possibly to initiation of protein synthesis with formylmethionine.
|
| |
Mol Cell Biol,
22,
5434-5442.
|
 |
|
|
|
|
 |
C.Mayer,
A.Stortchevoi,
C.Köhrer,
U.Varshney,
and
U.L.RajBhandary
(2001).
Initiator tRNA and its role in initiation of protein synthesis.
|
| |
Cold Spring Harb Symp Quant Biol,
66,
195-206.
|
 |
|
|
|
|
 |
J.Moser,
W.D.Schubert,
V.Beier,
I.Bringemeier,
D.Jahn,
and
D.W.Heinz
(2001).
V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis.
|
| |
EMBO J,
20,
6583-6590.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.D.Frankel
(2000).
Fitting peptides into the RNA world.
|
| |
Curr Opin Struct Biol,
10,
332-340.
|
 |
|
|
|
|
 |
G.R.Andersen,
L.Pedersen,
L.Valente,
I.Chatterjee,
T.G.Kinzy,
M.Kjeldgaard,
and
J.Nyborg
(2000).
Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Balpha.
|
| |
Mol Cell,
6,
1261-1266.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.S.Gabashvili,
R.K.Agrawal,
C.M.Spahn,
R.A.Grassucci,
D.I.Svergun,
J.Frank,
and
P.Penczek
(2000).
Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
|
| |
Cell,
100,
537-549.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Ibba,
and
D.Soll
(2000).
Aminoacyl-tRNA synthesis.
|
| |
Annu Rev Biochem,
69,
617-650.
|
 |
|
|
|
|
 |
S.Blanquet,
Y.Mechulam,
and
E.Schmitt
(2000).
The many routes of bacterial transfer RNAs after aminoacylation.
|
| |
Curr Opin Struct Biol,
10,
95.
|
 |
|
|
|
|
 |
S.Gite,
Y.Li,
V.Ramesh,
and
U.L.RajBhandary
(2000).
Escherichia coli methionyl-tRNA formyltransferase: role of amino acids conserved in the linker region and in the C-terminal domain on the specific recognition of the initiator tRNA.
|
| |
Biochemistry,
39,
2218-2226.
|
 |
|
|
|
|
 |
Y.Li,
W.B.Holmes,
D.R.Appling,
and
U.L.RajBhandary
(2000).
Initiation of protein synthesis in Saccharomyces cerevisiae mitochondria without formylation of the initiator tRNA.
|
| |
J Bacteriol,
182,
2886-2892.
|
 |
|
|
|
|
 |
C.H.Weber,
Y.S.Park,
S.Sanker,
C.Kent,
and
M.L.Ludwig
(1999).
A prototypical cytidylyltransferase: CTP:glycerol-3-phosphate cytidylyltransferase from bacillus subtilis.
|
| |
Structure,
7,
1113-1124.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.T.Newton,
M.Niemkiewicz,
R.Y.Lo,
and
D.Mangroo
(1999).
Recognition of the initiator tRNA by the Pseudomonas aeruginosa methionyl-tRNA formyltransferase: importance of the base-base mismatch at the end of the acceptor stem.
|
| |
FEMS Microbiol Lett,
178,
289-298.
|
 |
|
|
|
|
 |
M.Ibba,
and
D.Söll
(1999).
Quality control mechanisms during translation.
|
| |
Science,
286,
1893-1897.
|
 |
|
|
|
|
 |
P.J.Beuning,
and
K.Musier-Forsyth
(1999).
Transfer RNA recognition by aminoacyl-tRNA synthetases.
|
| |
Biopolymers,
52,
1.
|
 |
|
|
|
|
 |
S.Cusack
(1999).
RNA-protein complexes.
|
| |
Curr Opin Struct Biol,
9,
66-73.
|
 |
|
|
|
|
 |
V.Ramesh,
C.Mayer,
M.R.Dyson,
S.Gite,
and
U.L.RajBhandary
(1999).
Induced fit of a peptide loop of methionyl-tRNA formyltransferase triggered by the initiator tRNA substrate.
|
| |
Proc Natl Acad Sci U S A,
96,
875-880.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

