 |
PDBsum entry 2dps

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.2.6
- lysine/arginine leucyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
N-terminal L-lysyl-[protein] + L-leucyl-tRNA(Leu) = N-terminal L-leucyl-L-lysyl-[protein] + tRNA(Leu) + H(+)
|
|
2.
|
N-terminal L-arginyl-[protein] + L-leucyl-tRNA(Leu) = N-terminal L-leucyl-L-arginyl-[protein] + tRNA(Leu) + H(+)
|
|
 |
 |
 |
 |
 |
N-terminal L-lysyl-[protein]
|
+
|
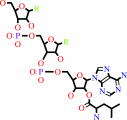 L-leucyl-tRNA(Leu)
L-leucyl-tRNA(Leu)
|
=
|
N-terminal L-leucyl-L-lysyl-[protein]
|
+
|
 tRNA(Leu)
tRNA(Leu)
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
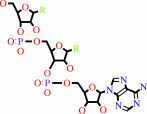
N-terminal L-arginyl-[protein]
|
+
|
 L-leucyl-tRNA(Leu)
L-leucyl-tRNA(Leu)
|
=
|
N-terminal L-leucyl-L-arginyl-[protein]
|
+
|
 tRNA(Leu)
tRNA(Leu)
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Monovalent cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
EMBO J
25:5942-5950
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of leucyl/phenylalanyl-tRNA-protein transferase and its complex with an aminoacyl-tRNA analog.
|
|
K.Suto,
Y.Shimizu,
K.Watanabe,
T.Ueda,
S.Fukai,
O.Nureki,
K.Tomita.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Eubacterial leucyl/phenylalanyl-tRNA protein transferase (L/F-transferase),
encoded by the aat gene, conjugates leucine or phenylalanine to the N-terminal
Arg or Lys residue of proteins, using Leu-tRNA(Leu) or Phe-tRNA(Phe) as a
substrate. The resulting N-terminal Leu or Phe acts as a degradation signal for
the ClpS-ClpAP-mediated N-end rule protein degradation pathway. Here, we present
the crystal structures of Escherichia coli L/F-transferase and its complex with
an aminoacyl-tRNA analog, puromycin. The C-terminal domain of L/F-transferase
consists of the GCN5-related N-acetyltransferase fold, commonly observed in the
acetyltransferase superfamily. The p-methoxybenzyl group of puromycin,
corresponding to the side chain of Leu or Phe of Leu-tRNA(Leu) or Phe-tRNA(Phe),
is accommodated in a highly hydrophobic pocket, with a shape and size suitable
for hydrophobic amino-acid residues lacking a branched beta-carbon, such as
leucine and phenylalanine. Structure-based mutagenesis of L/F-transferase
revealed its substrate specificity. Furthermore, we present a model of the
L/F-transferase complex with tRNA and substrate proteins bearing an N-terminal
Arg or Lys.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 Overall architecture of E. coli L/F-transferase. (A)
Stereo view of the E. coli L/F-transferase structure. The
NH[2]-terminal domain (residues 2–62) and the COOH-terminal
domain (residues 63–232) are colored blue and green,
respectively. The puromycin bound to the hydrophobic pocket is
colored yellow. (B) Topology diagram of L/F-transferase. The
rimmed elements in the COOH-terminal domain (  3– 3–
 5)
and ( 5)
and (  5– 5–
 12)
are common to the GNAT superfamily fold. The 12)
are common to the GNAT superfamily fold. The  -helices
and -helices
and  -strands
in the COOH-terminal domains are colored red and yellow,
respectively. (C) Comparison of the structures of E. coli
L/F-transferase (left), W. viridescens FemX (wvFemX; middle, PDB
accession number 1P4N; Biarrotte-Sorin et al, 2004) and S.
aureus FemA (saFemA; PDB accession number 1LRZ; Benson et al,
2002). The COOH-terminal domain of L/F-transferase is
topologically similar to the domain 2's of wvFemX and saFemA.
The conserved -strands
in the COOH-terminal domains are colored red and yellow,
respectively. (C) Comparison of the structures of E. coli
L/F-transferase (left), W. viridescens FemX (wvFemX; middle, PDB
accession number 1P4N; Biarrotte-Sorin et al, 2004) and S.
aureus FemA (saFemA; PDB accession number 1LRZ; Benson et al,
2002). The COOH-terminal domain of L/F-transferase is
topologically similar to the domain 2's of wvFemX and saFemA.
The conserved  -helices
and -helices
and  -strands
in L/F-transferase, wvFemX and saFemA, are colored red and
yellow, respectively. -strands
in L/F-transferase, wvFemX and saFemA, are colored red and
yellow, respectively.
|
 |
Figure 3.
Figure 3 Recognition of the puromycin by E. coli
L/F-transferase. (A) Chemical structure of puromycin (left) and
that of the 3'-ends of Leu-tRNA^Leu and Phe-tRNA^Phe (middle and
right, respectively). The amino-acid moiety and the base moiety
are colored pink and blue, respectively. (B) |Fo-Fc| omit map of
puromycin (contour level 3.0  ).
(C) Recognition of the p-methoxybenzyl group and the puromycin
base by the hydrophobic pocket, as shown by a surface model. (D)
Ribbon model of (C). The hydrophobic amino acid involved in the
recognition of the p-methoxybenzyl group and the base moiety of
puromycin are colored green and blue, respectively. (E) The
C-shaped edge of the hydrophobic pocket is composed of
continuous amino-acid residues (Gly155-Glu156-Ser157-Met158;
colored yellow and highlighted). The ).
(C) Recognition of the p-methoxybenzyl group and the puromycin
base by the hydrophobic pocket, as shown by a surface model. (D)
Ribbon model of (C). The hydrophobic amino acid involved in the
recognition of the p-methoxybenzyl group and the base moiety of
puromycin are colored green and blue, respectively. (E) The
C-shaped edge of the hydrophobic pocket is composed of
continuous amino-acid residues (Gly155-Glu156-Ser157-Met158;
colored yellow and highlighted). The  -, -,
 -
and -
and  -carbons
of puromycin are also shown. -carbons
of puromycin are also shown.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
EMBO J
(2006,
25,
5942-5950)
copyright 2006.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.Banerjee,
S.Chen,
K.Dare,
M.Gilreath,
M.Praetorius-Ibba,
M.Raina,
N.M.Reynolds,
T.Rogers,
H.Roy,
S.S.Yadavalli,
and
M.Ibba
(2010).
tRNAs: cellular barcodes for amino acids.
|
| |
FEBS Lett,
584,
387-395.
|
 |
|
|
|
|
 |
H.von Döhren
(2009).
Charged tRNAs charge into secondary metabolism.
|
| |
Nat Chem Biol,
5,
374-375.
|
 |
|
|
|
|
 |
M.Fonvielle,
M.Chemama,
R.Villet,
M.Lecerf,
A.Bouhss,
J.M.Valéry,
M.Ethève-Quelquejeu,
and
M.Arthur
(2009).
Aminoacyl-tRNA recognition by the FemXWv transferase for bacterial cell wall synthesis.
|
| |
Nucleic Acids Res,
37,
1589-1601.
|
 |
|
|
|
|
 |
R.L.Ninnis,
S.K.Spall,
G.H.Talbo,
K.N.Truscott,
and
D.A.Dougan
(2009).
Modification of PATase by L/F-transferase generates a ClpS-dependent N-end rule substrate in Escherichia coli.
|
| |
EMBO J,
28,
1732-1744.
|
 |
|
|
|
|
 |
J.L.Mainardi,
R.Villet,
T.D.Bugg,
C.Mayer,
and
M.Arthur
(2008).
Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria.
|
| |
FEMS Microbiol Rev,
32,
386-408.
|
 |
|
|
|
|
 |
L.M.Iyer,
A.M.Burroughs,
and
L.Aravind
(2008).
Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination.
|
| |
Biol Direct,
3,
45.
|
 |
|
|
|
|
 |
M.Taki,
H.Kuroiwa,
and
M.Sisido
(2008).
Chemoenzymatic transfer of fluorescent non-natural amino acids to the N terminus of a protein/peptide.
|
| |
Chembiochem,
9,
719-722.
|
 |
|
|
|
|
 |
Z.Xia,
A.Webster,
F.Du,
K.Piatkov,
M.Ghislain,
and
A.Varshavsky
(2008).
Substrate-binding Sites of UBR1, the Ubiquitin Ligase of the N-end Rule Pathway.
|
| |
J Biol Chem,
283,
24011-24028.
|
 |
|
|
|
|
 |
A.Mogk,
R.Schmidt,
and
B.Bukau
(2007).
The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies.
|
| |
Trends Cell Biol,
17,
165-172.
|
 |
|
|
|
|
 |
K.Watanabe,
Y.Toh,
K.Suto,
Y.Shimizu,
N.Oka,
T.Wada,
and
K.Tomita
(2007).
Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase.
|
| |
Nature,
449,
867-871.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Villet,
M.Fonvielle,
P.Busca,
M.Chemama,
A.P.Maillard,
J.E.Hugonnet,
L.Dubost,
A.Marie,
N.Josseaume,
S.Mesnage,
C.Mayer,
J.M.Valéry,
M.Ethève-Quelquejeu,
and
M.Arthur
(2007).
Idiosyncratic features in tRNAs participating in bacterial cell wall synthesis.
|
| |
Nucleic Acids Res,
35,
6870-6883.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

