 |
PDBsum entry 2edc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.8.1.5
- haloalkane dehalogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
1-haloalkane + H2O = a halide anion + a primary alcohol + H+
|
 |
 |
 |
 |
 |
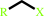 1-haloalkane
1-haloalkane
|
+
|
H2O
|
=
|
halide anion
|
+
|
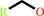 primary alcohol
primary alcohol
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
32:9031-9037
(1993)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallographic and fluorescence studies of the interaction of haloalkane dehalogenase with halide ions. Studies with halide compounds reveal a halide binding site in the active site.
|
|
K.H.Verschueren,
J.Kingma,
H.J.Rozeboom,
K.H.Kalk,
D.B.Janssen,
B.W.Dijkstra.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Haloalkane dehalogenase from Xanthobacter autotrophicus GJ10 catalyzes the
conversion of 1,2-dichloroethane to 2-chloroethanol and chloride without use of
oxygen or cofactors. The active site is situated in an internal cavity, which is
accessible from the solvent, even in the crystal. Crystal structures of the
dehalogenase enzyme complexed with iodoacetamide, chloroacetamide, iodide, and
chloride at pH 6.2 and 8.2 revealed a halide binding site between the ring NH's
of two tryptophan residues, Trp-125 and Trp-175, located in the active site. The
halide ion lies on the intersection of the planes of the rings of the
tryptophans. The binding of iodide and chloride to haloalkane dehalogenase
caused a strong decrease in protein fluorescence. The decrease could be fitted
to a modified form of the Stern-Volmer equation, indicating the presence of
fluorophors of different accessibilities. Halide binding was much stronger at pH
6.0 than at pH 8.2. Assuming ligand binding to Trp-125 and Trp-175 as the sole
cause of fluorescence quenching, dissociation constants at pH 6.0 with chloride
and iodide were calculated to be 0.49 +/- 0.04 and 0.074 +/- 0.007 mM,
respectively. Detailed structural investigation showed that the halide binding
site probably stabilizes the halide product as well as the negatively charged
transition state occurring during the formation of the covalent intermediate.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.A.Robertson,
G.K.Schroeder,
Z.Jin,
K.A.Johnson,
and
C.P.Whitman
(2009).
Pre-steady-state kinetic analysis of cis-3-chloroacrylic acid dehalogenase: analysis and implications.
|
| |
Biochemistry,
48,
11737-11744.
|
 |
|
|
|
|
 |
Y.R.Gokarn,
R.M.Fesinmeyer,
A.Saluja,
S.Cao,
J.Dankberg,
A.Goetze,
R.L.Remmele,
L.O.Narhi,
and
D.N.Brems
(2009).
Ion-specific modulation of protein interactions: anion-induced, reversible oligomerization of a fusion protein.
|
| |
Protein Sci,
18,
169-179.
|
 |
|
|
|
|
 |
E.Chovancová,
J.Kosinski,
J.M.Bujnicki,
and
J.Damborský
(2007).
Phylogenetic analysis of haloalkane dehalogenases.
|
| |
Proteins,
67,
305-316.
|
 |
|
|
|
|
 |
M.Petrek,
M.Otyepka,
P.Banás,
P.Kosinová,
J.Koca,
and
J.Damborský
(2006).
CAVER: a new tool to explore routes from protein clefts, pockets and cavities.
|
| |
BMC Bioinformatics,
7,
316.
|
 |
|
|
|
|
 |
C.Colin,
C.Leblanc,
G.Michel,
E.Wagner,
E.Leize-Wagner,
A.Van Dorsselaer,
and
P.Potin
(2005).
Vanadium-dependent iodoperoxidases in Laminaria digitata, a novel biochemical function diverging from brown algal bromoperoxidases.
|
| |
J Biol Inorg Chem,
10,
156-166.
|
 |
|
|
|
|
 |
E.Szolajska,
J.Poznanski,
M.L.Ferber,
J.Michalik,
E.Gout,
P.Fender,
I.Bailly,
B.Dublet,
and
J.Chroboczek
(2004).
Poneratoxin, a neurotoxin from ant venom. Structure and expression in insect cells and construction of a bio-insecticide.
|
| |
Eur J Biochem,
271,
2127-2136.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Ohshiro,
J.Littlechild,
E.Garcia-Rodriguez,
M.N.Isupov,
Y.Iida,
T.Kobayashi,
and
Y.Izumi
(2004).
Modification of halogen specificity of a vanadium-dependent bromoperoxidase.
|
| |
Protein Sci,
13,
1566-1571.
|
 |
|
|
|
|
 |
G.J.Poelarends,
R.Saunier,
and
D.B.Janssen
(2001).
trans-3-Chloroacrylic acid dehalogenase from Pseudomonas pavonaceae 170 shares structural and mechanistic similarities with 4-oxalocrotonate tautomerase.
|
| |
J Bacteriol,
183,
4269-4277.
|
 |
|
|
|
|
 |
U.Kaulmann,
S.R.Kaschabek,
and
M.Schlömann
(2001).
Mechanism of chloride elimination from 3-chloro- and 2,4-dichloro-cis,cis-muconate: new insight obtained from analysis of muconate cycloisomerase variant CatB-K169A.
|
| |
J Bacteriol,
183,
4551-4561.
|
 |
|
|
|
|
 |
M.E.Walsh,
P.Kyritsis,
N.A.Eady,
H.A.Hill,
and
L.L.Wong
(2000).
Catalytic reductive dehalogenation of hexachloroethane by molecular variants of cytochrome P450cam (CYP101).
|
| |
Eur J Biochem,
267,
5815-5820.
|
 |
|
|
|
|
 |
T.C.Bruice,
and
S.J.Benkovic
(2000).
Chemical basis for enzyme catalysis.
|
| |
Biochemistry,
39,
6267-6274.
|
 |
|
|
|
|
 |
I.S.Ridder,
H.J.Rozeboom,
and
B.W.Dijkstra
(1999).
Haloalkane dehalogenase from Xanthobacter autotrophicus GJ10 refined at 1.15 A resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
1273-1290.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.S.Ridder,
H.J.Rozeboom,
K.H.Kalk,
and
B.W.Dijkstra
(1999).
Crystal structures of intermediates in the dehalogenation of haloalkanoates by L-2-haloacid dehalogenase.
|
| |
J Biol Chem,
274,
30672-30678.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.F.Schindler,
P.A.Naranjo,
D.A.Honaberger,
C.H.Chang,
J.R.Brainard,
L.A.Vanderberg,
and
C.J.Unkefer
(1999).
Haloalkane dehalogenases: steady-state kinetics and halide inhibition.
|
| |
Biochemistry,
38,
5772-5778.
|
 |
|
|
|
|
 |
W.Chen,
F.Brühlmann,
R.D.Richins,
and
A.Mulchandani
(1999).
Engineering of improved microbes and enzymes for bioremediation.
|
| |
Curr Opin Biotechnol,
10,
137-141.
|
 |
|
|
|
|
 |
A.Butler
(1998).
Vanadium haloperoxidases.
|
| |
Curr Opin Chem Biol,
2,
279-285.
|
 |
|
|
|
|
 |
G.H.Krooshof,
I.S.Ridder,
A.W.Tepper,
G.J.Vos,
H.J.Rozeboom,
K.H.Kalk,
B.W.Dijkstra,
and
D.B.Janssen
(1998).
Kinetic analysis and X-ray structure of haloalkane dehalogenase with a modified halide-binding site.
|
| |
Biochemistry,
37,
15013-15023.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.R.Albani
(1998).
Dynamics of the Lens culinaris agglutinin-lactotransferrin and serotransferrin complexes, followed by fluorescence intensity quenching of fluorescein (FITC) with iodide and temperature.
|
| |
Biochim Biophys Acta,
1425,
405-410.
|
 |
|
|
|
|
 |
L.T.Laughlin,
H.F.Tzeng,
S.Lin,
and
R.N.Armstrong
(1998).
Mechanism of microsomal epoxide hydrolase. Semifunctional site-specific mutants affecting the alkylation half-reaction.
|
| |
Biochemistry,
37,
2897-2904.
|
 |
|
|
|
|
 |
P.Holloway,
K.L.Knoke,
J.T.Trevors,
and
H.Lee
(1998).
Alteration of the substrate range of haloalkane dehalogenase by site-directed mutagenesis.
|
| |
Biotechnol Bioeng,
59,
520-523.
|
 |
|
|
|
|
 |
F.C.Lightstone,
Y.J.Zheng,
A.H.Maulitz,
and
T.C.Bruice
(1997).
Non-enzymatic and enzymatic hydrolysis of alkyl halides: a haloalkane dehalogenation enzyme evolved to stabilize the gas-phase transition state of an SN2 displacement reaction.
|
| |
Proc Natl Acad Sci U S A,
94,
8417-8420.
|
 |
|
|
|
|
 |
G.H.Krooshof,
E.M.Kwant,
J.Damborský,
J.Koca,
and
D.B.Janssen
(1997).
Repositioning the catalytic triad aspartic acid of haloalkane dehalogenase: effects on stability, kinetics, and structure.
|
| |
Biochemistry,
36,
9571-9580.
|
 |
|
|
|
|
 |
A.Messerschmidt,
and
R.Wever
(1996).
X-ray structure of a vanadium-containing enzyme: chloroperoxidase from the fungus Curvularia inaequalis.
|
| |
Proc Natl Acad Sci U S A,
93,
392-396.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Yang,
R.Q.Liu,
K.L.Taylor,
H.Xiang,
J.Price,
and
D.Dunaway-Mariano
(1996).
Identification of active site residues essential to 4-chlorobenzoyl-coenzyme A dehalogenase catalysis by chemical modification and site directed mutagenesis.
|
| |
Biochemistry,
35,
10879-10885.
|
 |
|
|
|
|
 |
J.I.Manchester,
and
R.L.Ornstein
(1996).
Rational approach to improving reductive catalysis by cytochrome P450cam.
|
| |
Biochimie,
78,
714-722.
|
 |
|
|
|
|
 |
J.P.Schanstra,
and
D.B.Janssen
(1996).
Kinetics of halide release of haloalkane dehalogenase: evidence for a slow conformational change.
|
| |
Biochemistry,
35,
5624-5632.
|
 |
|
|
|
|
 |
J.P.Schanstra,
I.S.Ridder,
G.J.Heimeriks,
R.Rink,
G.J.Poelarends,
K.H.Kalk,
B.W.Dijkstra,
and
D.B.Janssen
(1996).
Kinetic characterization and X-ray structure of a mutant of haloalkane dehalogenase with higher catalytic activity and modified substrate range.
|
| |
Biochemistry,
35,
13186-13195.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.P.Schanstra,
J.Kingma,
and
D.B.Janssen
(1996).
Specificity and kinetics of haloalkane dehalogenase.
|
| |
J Biol Chem,
271,
14747-14753.
|
 |
|
|
|
|
 |
Y.Chen,
J.Inobe,
V.K.Kuchroo,
J.L.Baron,
C.A.Janeway,
and
H.L.Weiner
(1996).
Oral tolerance in myelin basic protein T-cell receptor transgenic mice: suppression of autoimmune encephalomyelitis and dose-dependent induction of regulatory cells.
|
| |
Proc Natl Acad Sci U S A,
93,
388-391.
|
 |
|
|
|
|
 |
C.Kennes,
F.Pries,
G.H.Krooshof,
E.Bokma,
J.Kingma,
and
D.B.Janssen
(1995).
Replacement of tryptophan residues in haloalkane dehalogenase reduces halide binding and catalytic activity.
|
| |
Eur J Biochem,
228,
403-407.
|
 |
|
|
|
|
 |
D.B.Janssen,
J.R.van der Ploeg,
and
F.Pries
(1995).
Genetic adaptation of bacteria to halogenated aliphatic compounds.
|
| |
Environ Health Perspect,
103,
29-32.
|
 |
|
|
|
|
 |
F.Pries,
J.Kingma,
G.H.Krooshof,
C.M.Jeronimus-Stratingh,
A.P.Bruins,
and
D.B.Janssen
(1995).
Histidine 289 is essential for hydrolysis of the alkyl-enzyme intermediate of haloalkane dehalogenase.
|
| |
J Biol Chem,
270,
10405-10411.
|
 |
|
|
|
|
 |
D.B.Janssen,
J.R.van der Ploeg,
and
F.Pries
(1994).
Genetics and biochemistry of 1,2-dichloroethane degradation.
|
| |
Biodegradation,
5,
249-257.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

