 |
PDBsum entry 1be0

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.8.1.5
- haloalkane dehalogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
1-haloalkane + H2O = a halide anion + a primary alcohol + H+
|
 |
 |
 |
 |
 |
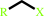 1-haloalkane
1-haloalkane
|
+
|
H2O
|
=
|
halide anion
|
+
|
primary alcohol
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
37:15013-15023
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Kinetic analysis and X-ray structure of haloalkane dehalogenase with a modified halide-binding site.
|
|
G.H.Krooshof,
I.S.Ridder,
A.W.Tepper,
G.J.Vos,
H.J.Rozeboom,
K.H.Kalk,
B.W.Dijkstra,
D.B.Janssen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Haloalkane dehalogenase (DhlA) catalyzes the hydrolysis of haloalkanes via an
alkyl-enzyme intermediate. Trp175 forms a halogen/halide-binding site in the
active-site cavity together with Trp125. To get more insight in the role of
Trp175 in DhlA, we mutated residue 175 and explored the kinetics and X-ray
structure of the Trp175Tyr enzyme. The mutagenesis study indicated that an
aromatic residue at position 175 is important for the catalytic performance of
DhlA. Pre-steady-state kinetic analysis of Trp175Tyr-DhlA showed that the
observed 6-fold increase of the Km for 1,2-dibromoethane (DBE) results from
reduced rates of both DBE binding and cleavage of the carbon-bromine bond.
Furthermore, the enzyme isomerization preceding bromide release became 4-fold
faster in the mutant enzyme. As a result, the rate of hydrolysis of the
alkyl-enzyme intermediate became the main determinant of the kcat for DBE, which
was 2-fold higher than the wild-type kcat. The X-ray structure of the mutant
enzyme at pH 6 showed that the backbone structure of the enzyme remains intact
and that the tyrosine side chain lies in the same plane as Trp175 in the
wild-type enzyme. The Clalpha-stabilizing aromatic rings of Tyr175 and Trp125
are 0.7 A further apart and due to the smaller size of the mutated residue, the
volume of the cavity has increased by one-fifth. X-ray structures of mutant and
wild-type enzyme at pH 5 demonstrated that the Tyr175 side chain rotated away
upon binding of an acetic acid molecule, leaving one of its oxygen atoms
hydrogen bonded to the indole nitrogen of Trp125 only. These structural changes
indicate a weakened interaction between residue 175 and the halogen atom or
halide ion in the active site and help to explain the kinetic changes induced by
the Trp175Tyr mutation.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.Valero,
L.Song,
J.Gao,
and
D.G.Truhlar
(2009).
Perspective on Diabatic Models of Chemical Reactivity as Illustrated by the Gas-Phase S(N)2 Reaction of Acetate Ion with 1,2-Dichloroethane.
|
| |
J Chem Theory Comput,
5,
1.
|
 |
|
|
|
|
 |
M.Petrek,
M.Otyepka,
P.Banás,
P.Kosinová,
J.Koca,
and
J.Damborský
(2006).
CAVER: a new tool to explore routes from protein clefts, pockets and cavities.
|
| |
BMC Bioinformatics,
7,
316.
|
 |
|
|
|
|
 |
D.B.Janssen
(2004).
Evolving haloalkane dehalogenases.
|
| |
Curr Opin Chem Biol,
8,
150-159.
|
 |
|
|
|
|
 |
Z.Prokop,
M.Monincová,
R.Chaloupková,
M.Klvana,
Y.Nagata,
D.B.Janssen,
and
J.Damborský
(2003).
Catalytic mechanism of the maloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26.
|
| |
J Biol Chem,
278,
45094-45100.
|
 |
|
|
|
|
 |
G.J.Poelarends,
R.Saunier,
and
D.B.Janssen
(2001).
trans-3-Chloroacrylic acid dehalogenase from Pseudomonas pavonaceae 170 shares structural and mechanistic similarities with 4-oxalocrotonate tautomerase.
|
| |
J Bacteriol,
183,
4269-4277.
|
 |
|
|
|
|
 |
E.Y.Lau,
K.Kahn,
P.A.Bash,
and
T.C.Bruice
(2000).
The importance of reactant positioning in enzyme catalysis: a hybrid quantum mechanics/molecular mechanics study of a haloalkane dehalogenase.
|
| |
Proc Natl Acad Sci U S A,
97,
9937-9942.
|
 |
|
|
|
|
 |
M.E.Walsh,
P.Kyritsis,
N.A.Eady,
H.A.Hill,
and
L.L.Wong
(2000).
Catalytic reductive dehalogenation of hexachloroethane by molecular variants of cytochrome P450cam (CYP101).
|
| |
Eur J Biochem,
267,
5815-5820.
|
 |
|
|
|
|
 |
G.H.Krooshof,
R.Floris,
A.W.Tepper,
and
D.B.Janssen
(1999).
Thermodynamic analysis of halide binding to haloalkane dehalogenase suggests the occurrence of large conformational changes.
|
| |
Protein Sci,
8,
355-360.
|
 |
|
|
|
|
 |
I.S.Ridder,
H.J.Rozeboom,
and
B.W.Dijkstra
(1999).
Haloalkane dehalogenase from Xanthobacter autotrophicus GJ10 refined at 1.15 A resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
1273-1290.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.S.Ridder,
H.J.Rozeboom,
K.H.Kalk,
and
B.W.Dijkstra
(1999).
Crystal structures of intermediates in the dehalogenation of haloalkanoates by L-2-haloacid dehalogenase.
|
| |
J Biol Chem,
274,
30672-30678.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.F.Schindler,
P.A.Naranjo,
D.A.Honaberger,
C.H.Chang,
J.R.Brainard,
L.A.Vanderberg,
and
C.J.Unkefer
(1999).
Haloalkane dehalogenases: steady-state kinetics and halide inhibition.
|
| |
Biochemistry,
38,
5772-5778.
|
 |
|
|
|
|
 |
J.Newman,
T.S.Peat,
R.Richard,
L.Kan,
P.E.Swanson,
J.A.Affholter,
I.H.Holmes,
J.F.Schindler,
C.J.Unkefer,
and
T.C.Terwilliger
(1999).
Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
|
| |
Biochemistry,
38,
16105-16114.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.E.Swanson
(1999).
Dehalogenases applied to industrial-scale biocatalysis.
|
| |
Curr Opin Biotechnol,
10,
365-369.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

