 |
PDBsum entry 2c1c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Structural basis of the resistance of an insect carboxypeptidase to plant protease inhibitors
|
|
Structure:
|
 |
Carboxypeptidase b. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Helicoverpa zea. Corn earworm. Organism_taxid: 7113. Tissue: epithelium. Expressed in: pichia pastoris. Expression_system_taxid: 4922.
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.215
|
R-free:
|
0.292
|
|
|
Authors:
|
 |
A.Bayes,M.Comellas-Bigler,M.Rodriguez De La Vega,K.Maskos,W.Bode, F.X.Aviles,M.A.Jongsma,J.Beekwilder,J.Vendrell
|
Key ref:

|
 |
A.Bayés
et al.
(2005).
Structural basis of the resistance of an insect carboxypeptidase to plant protease inhibitors.
Proc Natl Acad Sci U S A,
102,
16602-16607.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
12-Sep-05
|
Release date:
|
19-Oct-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q3T905
(CBPB_HELZE) -
Carboxypeptidase B from Helicoverpa zea
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
429 a.a.
312 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.4.17.2
- carboxypeptidase B.
|
|
 |
 |
 |
 |
 |
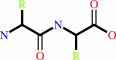
Reaction:
|
 |
Peptidyl-L-lysine(or L-arginine) + H(2)O = peptide + L-lysine(or L- arginine)
|
 |
 |
 |
 |
 |
|
+
|
|
=
|
|
+
|
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
102:16602-16607
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis of the resistance of an insect carboxypeptidase to plant protease inhibitors.
|
|
A.Bayés,
M.Comellas-Bigler,
M.Rodríguez de la Vega,
K.Maskos,
W.Bode,
F.X.Aviles,
M.A.Jongsma,
J.Beekwilder,
J.Vendrell.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Corn earworm (Helicoverpa zea), also called tomato fruitworm, is a common pest
of many Solanaceous plants. This insect is known to adapt to the ingestion of
plant serine protease inhibitors by using digestive proteases that are
insensitive to inhibition. We have now identified a B-type carboxypeptidase of
H. zea (CPBHz) insensitive to potato carboxypeptidase inhibitor (PCI) in corn
earworm. To elucidate the structural features leading to the adaptation of the
insect enzyme, the crystal structure of the recombinant CPBHz protein was
determined by x-ray diffraction. CPBHz is a member of the A/B subfamily of
metallocarboxypeptidases, which displays the characteristic
metallocarboxypeptidase alpha/beta-hydrolase fold, and does not differ
essentially from the previously described Helicoverpa armigera CPA, which is
very sensitive to PCI. The data provide structural insight into several
functional properties of CPBHz. The high selectivity shown by CPBHz for
C-terminal lysine residues is due to residue changes in the S1' substrate
specificity pocket that render it unable to accommodate the side chain of an
arginine. The insensitivity of CPBHz to plant inhibitors is explained by the
exceptional positioning of two of the main regions that stabilize other
carboxypeptidase-PCI complexes, the beta8-alpha9 loop, and alpha7 together with
the alpha7-alpha8 loop. The rearrangement of these two regions leads to a
displacement of the active-site entrance that impairs the proper interaction
with PCI. This report explains a crystal structure of an insect protease and its
adaptation to defensive plant protease inhibitors.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Stereo ribbon plot representation of CPBHz. The two
histidines and the glutamic acid residues coordinating the zinc
atom are shown as stick models in green, the zinc atom is shown
as a silver sphere, and the single disulfide bridge present in
the structure is shown in blue. The polypeptide fold consists of
a core of eight twisted  -sheets surrounded by
eight -sheets surrounded by
eight  -helices. Figure
generated with MOLSCRIPT (31) and RASTER 3D (32). -helices. Figure
generated with MOLSCRIPT (31) and RASTER 3D (32).
|
 |
Figure 3.
Fig. 3. Stereoview of a structural superimposition of H.
zea carboxypeptidase B in orange, human carboxypeptidase B in
green, and H. armigera carboxypeptidase A, in purple. Trp-277A
of CPBHz and residues Trp-277B and Tyr-277 in human CPAHa and
CPBh, respectively, are represented as stick models; the arrow
indicates the entrance to the active-site cleft. The zinc ion is
shown in silver. The orientation of the molecule is the same as
in Fig. 1.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.M.Dunse,
Q.Kaas,
R.F.Guarino,
P.A.Barton,
D.J.Craik,
and
M.A.Anderson
(2010).
Molecular basis for the resistance of an insect chymotrypsin to a potato type II proteinase inhibitor.
|
| |
Proc Natl Acad Sci U S A,
107,
15016-15021.
|
 |
|
|
|
|
 |
D.Fernández,
F.X.Avilés,
and
J.Vendrell
(2009).
Aromatic organic compounds as scaffolds for metallocarboxypeptidase inhibitor design.
|
| |
Chem Biol Drug Des,
73,
75-82.
|
 |
|
|
|
|
 |
G.A.Howe,
and
G.Jander
(2008).
Plant immunity to insect herbivores.
|
| |
Annu Rev Plant Biol,
59,
41-66.
|
 |
|
|
|
|
 |
J.A.Zavala,
A.P.Giri,
M.A.Jongsma,
and
I.T.Baldwin
(2008).
Digestive duet: midgut digestive proteinases of Manduca sexta ingesting Nicotiana attenuata with manipulated trypsin proteinase inhibitor expression.
|
| |
PLoS ONE,
3,
e2008.
|
 |
|
|
|
|
 |
Y.D.Koo,
J.E.Ahn,
R.A.Salzman,
J.Moon,
Y.H.Chi,
D.J.Yun,
S.Y.Lee,
H.Koiwa,
and
K.Zhu-Salzman
(2008).
Functional expression of an insect cathepsin B-like counter-defence protein.
|
| |
Insect Mol Biol,
17,
235-245.
|
 |
|
|
|
|
 |
D.Fernández,
J.Vendrell,
F.X.Avilés,
and
J.Fernández-Recio
(2007).
Structural and functional characterization of binding sites in metallocarboxypeptidases based on Optimal Docking Area analysis.
|
| |
Proteins,
68,
131-144.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

