 |
PDBsum entry 2agd

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of human m340h-beta-1,4-galactosyltransferase- i(m340h-b4gal-t1) in complex with glcnac-beta1,4-man-alpha1,3-man- beta-or
|
|
Structure:
|
 |
Beta-1,4-galactosyltransferase 1. Chain: a, b, c. Fragment: catalytic domain, residues 126-398. Synonym: beta-1,4-galtase 1. Beta4gal-t1. B4gal-t1. Udp- galactose:beta-n-acetylglucosamine beta-1,4-galactosyltransferase 1. Engineered: yes. Mutation: yes. Other_details: n-acetyllactosamine synthase part
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: b4galt1, ggtb2. Expressed in: escherichia coli bl21. Expression_system_taxid: 511693. Other_details: n-terminal carries t7 tag of 11 amino acids followed by gly, ser & ala
|
|
Resolution:
|
 |
|
1.90Å
|
R-factor:
|
0.202
|
R-free:
|
0.228
|
|
|
Authors:
|
 |
V.Ramasamy,B.Ramakrishnan,E.Boeggeman,D.M.Ratner,P.H.Seeberger, P.K.Qasba
|
Key ref:

|
 |
V.Ramasamy
et al.
(2005).
Oligosaccharide preferences of beta1,4-galactosyltransferase-I: crystal structures of Met340His mutant of human beta1,4-galactosyltransferase-I with a pentasaccharide and trisaccharides of the N-glycan moiety.
J Mol Biol,
353,
53-67.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
26-Jul-05
|
Release date:
|
04-Oct-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P15291
(B4GT1_HUMAN) -
Beta-1,4-galactosyltransferase 1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
398 a.a.
273 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 3 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.4.1.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.4.1.22
- lactose synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-glucose + UDP-alpha-D-galactose = lactose + UDP + H+
|
 |
 |
 |
 |
 |
D-glucose
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
UDP-alpha-D-galactose
|
=
|
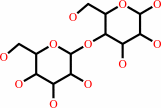 lactose
lactose
|
+
|
 UDP
UDP
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 78.12% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.2.4.1.275
- neolactotriaosylceramide beta-1,4-galactosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a beta-D-GlcNAc-(1->3)-beta-D-Gal-(1->4)-beta-D-Glc-(1<->1)- Cer(d18:1(4E)) + UDP-alpha-D-galactose = a neolactoside nLc4Cer(d18:1(4E)) + UDP + H+
|
 |
 |
 |
 |
 |
beta-D-GlcNAc-(1->3)-beta-D-Gal-(1->4)-beta-D-Glc-(1<->1)- Cer(d18:1(4E))
|
+
|
UDP-alpha-D-galactose
|
=
|
neolactoside nLc4Cer(d18:1(4E))
|
+
|
 UDP
UDP
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 78.12% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.2.4.1.38
- beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an N-acetyl-beta-D-glucosaminyl derivative + UDP-alpha-D-galactose = a beta-D-galactosyl-(1->4)-N-acetyl-beta-D-glucosaminyl derivative + UDP + H+
|
 |
 |
 |
 |
 |
N-acetyl-beta-D-glucosaminyl derivative
|
+
|
UDP-alpha-D-galactose
|
=
|
beta-D-galactosyl-(1->4)-N-acetyl-beta-D-glucosaminyl derivative
|
+
|
 UDP
UDP
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 78.12% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
E.C.2.4.1.90
- N-acetyllactosamine synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-acetyl-D-glucosamine + UDP-alpha-D-galactose = beta-D-galactosyl- (1->4)-N-acetyl-D-glucosamine + UDP + H+
|
 |
 |
 |
 |
 |
N-acetyl-D-glucosamine
Bound ligand (Het Group name = )
matches with 93.33% similarity
|
+
|
UDP-alpha-D-galactose
|
=
|
beta-D-galactosyl- (1->4)-N-acetyl-D-glucosamine
|
+
|
 UDP
UDP
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 78.12% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
353:53-67
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Oligosaccharide preferences of beta1,4-galactosyltransferase-I: crystal structures of Met340His mutant of human beta1,4-galactosyltransferase-I with a pentasaccharide and trisaccharides of the N-glycan moiety.
|
|
V.Ramasamy,
B.Ramakrishnan,
E.Boeggeman,
D.M.Ratner,
P.H.Seeberger,
P.K.Qasba.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
beta-1,4-Galactosyltransferase-I (beta4Gal-T1) transfers galactose from
UDP-galactose to N-acetylglucosamine (GlcNAc) residues of the branched N-linked
oligosaccharide chains of glycoproteins. In an N-linked biantennary
oligosaccharide chain, one antenna is attached to the 3-hydroxyl-(1,3-arm), and
the other to the 6-hydroxyl-(1,6-arm) group of mannose, which is beta-1,4-linked
to an N-linked chitobiose, attached to the aspargine residue of a protein. For a
better understanding of the branch specificity of beta4Gal-T1 towards the GlcNAc
residues of N-glycans, we have carried out kinetic and crystallographic studies
with the wild-type human beta4Gal-T1 (h-beta4Gal-T1) and the mutant
Met340His-beta4Gal-T1 (h-M340H-beta4Gal-T1) in complex with a GlcNAc-containing
pentasaccharide and several GlcNAc-containing trisaccharides present in
N-glycans. The oligosaccharides used were: pentasaccharide
GlcNAcbeta1,2-Manalpha1,6 (GlcNAcbeta1,2-Manalpha1,3)Man; the 1,6-arm
trisaccharide, GlcNAcbeta1,2-Manalpha1,6-Manbeta-OR (1,2-1,6-arm); the 1,3-arm
trisaccharides, GlcNAcbeta1,2-Manalpha1,3-Manbeta-OR (1,2-1,3-arm) and
GlcNAcbeta1,4-Manalpha1,3-Manbeta-OR (1,4-1,3-arm); and the trisaccharide
GlcNAcbeta1,4-GlcNAcbeta1,4-GlcNAc (chitotriose). With the wild-type
h-beta4Gal-T1, the K(m) of 1,2-1,6-arm is approximately tenfold lower than for
1,2-1,3-arm and 1,4-1,3-arm, and 22-fold lower than for chitotriose. Crystal
structures of h-M340H-beta4Gal-T1 in complex with the pentasaccharide and
various trisaccharides at 1.9-2.0A resolution showed that beta4Gal-T1 is in a
closed conformation with the oligosaccharide bound to the enzyme, and the
1,2-1,6-arm trisaccharide makes the maximum number of interactions with the
enzyme, which is in concurrence with the lowest K(m) for the trisaccharide.
Present studies suggest that beta4Gal-T1 interacts preferentially with the
1,2-1,6-arm trisaccharide rather than with the 1,2-1,3-arm or 1,4-1,3-arm of a
bi- or tri-antennary oligosaccharide chain of N-glycan.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. A depiction of complex penta-antennary N-glycan
structure. The pentasaccharide and the trisaccharides of the
N-glycan moiety, used for the kinetic and crystallographic
studies, are highlighted by colored arrows.
GlcNAcb1,2-Mana1,6(GlcNAcb1,2-Mana1,3)-Man pentasaccharide (blue
and green arrows); GlcNAcb1,2-Mana1,6-Manb-OR (1,2-1,6-arm)
(blue arrows); GlcNAcb1,2-Mana1,3-Manb-OR (1,2-1,3-arm) (green
arrows); GlcNAcb1,4-Man a1,3-Manb-OR (1,4-1,3-arm) (purple
arrows).
|
 |
Figure 2.
Figure 2. Effects of varying the oligosaccharide acceptor
substrate concentration on the initial rate of galactose
transfer (n) by the h-b4Gal-T1. Chitobiose (GlcNAcb1,4-GlcNAc)
and chitotriose GlcNAcb1,4-GlcNAcb1,4-GlcNAc (0M),
GlcNAcb1,2-Man (  triangle,
open ), GlcNAcb1,2-Mana1,6-Manb-OR (sB),
GlcNAcb1,2-Mana1,3-Manb-OR ( triangle,
open ), GlcNAcb1,2-Mana1,6-Manb-OR (sB),
GlcNAcb1,2-Mana1,3-Manb-OR (  open
), GlcNAcb1,4-Mana1,3-Manb-OR ( open
), GlcNAcb1,4-Mana1,3-Manb-OR (  triangle,
filled ), pentasaccharide
GlcNAcb1,2-Mana1,6(GlcNAcb1,2-Mana1,3)Man( triangle,
filled ), pentasaccharide
GlcNAcb1,2-Mana1,6(GlcNAcb1,2-Mana1,3)Man(  ). ).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2005,
353,
53-67)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.R.Brown,
F.Yang,
A.Sinha,
B.Ramakrishnan,
Y.Tor,
P.K.Qasba,
and
J.D.Esko
(2009).
Deoxygenated Disaccharide Analogs as Specific Inhibitors of {beta}1-4-Galactosyltransferase 1 and Selectin-mediated Tumor Metastasis.
|
| |
J Biol Chem,
284,
4952-4959.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Bojarová,
K.Krenek,
K.Wetjen,
K.Adamiak,
H.Pelantová,
K.Bezouska,
L.Elling,
and
V.Kren
(2009).
Synthesis of LacdiNAc-terminated glycoconjugates by mutant galactosyltransferase--a way to new glycodrugs and materials.
|
| |
Glycobiology,
19,
509-517.
|
 |
|
|
|
|
 |
T.Okada,
H.Ihara,
R.Ito,
N.Taniguchi,
and
Y.Ikeda
(2009).
Bidirectional N-acetylglucosamine transfer mediated by beta-1,4-N-acetylglucosaminyltransferase III.
|
| |
Glycobiology,
19,
368-374.
|
 |
|
|
|
|
 |
L.L.Lairson,
B.Henrissat,
G.J.Davies,
and
S.G.Withers
(2008).
Glycosyltransferases: structures, functions, and mechanisms.
|
| |
Annu Rev Biochem,
77,
521-555.
|
 |
|
|
|
|
 |
P.K.Qasba,
B.Ramakrishnan,
and
E.Boeggeman
(2008).
Structure and function of beta -1,4-galactosyltransferase.
|
| |
Curr Drug Targets,
9,
292-309.
|
 |
|
|
|
|
 |
I.Brockhausen,
M.Benn,
S.Bhat,
S.Marone,
J.G.Riley,
P.Montoya-Peleaz,
J.Z.Vlahakis,
H.Paulsen,
J.S.Schutzbach,
and
W.A.Szarek
(2006).
UDP-Gal: GlcNAc-R beta1,4-galactosyltransferase--a target enzyme for drug design. Acceptor specificity and inhibition of the enzyme.
|
| |
Glycoconj J,
23,
525-541.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

