 |
PDBsum entry 2yid
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of the suca domain of mycobacterium smegmatis alpha- ketoglutarate decarboxylase in complex with the enamine-thdp intermediate
|
|
Structure:
|
 |
2-oxoglutarate decarboxylase. Chain: a, b, c, d. Fragment: residues 361-1227. Synonym: 2-oxoglutarate carboxy-lyase, alpha-ketoglutarate decarboxylase, kdh. Engineered: yes
|
|
Source:
|
 |
Mycobacterium smegmatis. Organism_taxid: 1772. Strain: mc2155. Expressed in: escherichia coli. Expression_system_taxid: 469008. Expression_system_variant: plyss.
|
|
Resolution:
|
 |
|
2.25Å
|
R-factor:
|
0.194
|
R-free:
|
0.223
|
|
|
Authors:
|
 |
T.Wagner,M.Bellinzoni,A.M.Wehenkel,H.M.O'Hare,P.M.Alzari
|
|
Key ref:
|
 |
T.Wagner
et al.
(2011).
Functional plasticity and allosteric regulation of α-ketoglutarate decarboxylase in central mycobacterial metabolism.
Chem Biol,
18,
1011-1020.
PubMed id:
|
 |
|
Date:
|
 |
|
11-May-11
|
Release date:
|
15-Jun-11
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
A0R2B1
(KGD_MYCS2) -
Multifunctional 2-oxoglutarate metabolism enzyme from Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1227 a.a.
844 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.2.4.2
- oxoglutarate dehydrogenase (succinyl-transferring).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Oxo-acid dehydrogenase complexes
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N6-[(R)-lipoyl]-L-lysyl-[protein] + 2-oxoglutarate + H+ = N6-[(R)- S(8)-succinyldihydrolipoyl]-L-lysyl-[protein] + CO2
|
 |
 |
 |
 |
 |
N(6)-[(R)-lipoyl]-L-lysyl-[protein]
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
+
|
H(+)
|
=
|
N(6)-[(R)- S(8)-succinyldihydrolipoyl]-L-lysyl-[protein]
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Thiamine diphosphate
|
 |
 |
 |
 |
 |
Thiamine diphosphate
Bound ligand (Het Group name =
TD7)
matches with 78.79% similarity
|
|
 |
 |
Enzyme class 3:
|
 |
E.C.2.2.1.5
- 2-hydroxy-3-oxoadipate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
glyoxylate + 2-oxoglutarate + H+ = 2-hydroxy-3-oxoadipate + CO2
|
 |
 |
 |
 |
 |
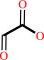 glyoxylate
glyoxylate
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
+
|
H(+)
|
=
|
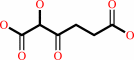 2-hydroxy-3-oxoadipate
2-hydroxy-3-oxoadipate
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Thiamine diphosphate
|
 |
 |
 |
 |
 |
Thiamine diphosphate
Bound ligand (Het Group name =
TD7)
matches with 78.79% similarity
|
|
 |
 |
Enzyme class 4:
|
 |
E.C.2.3.1.61
- dihydrolipoyllysine-residue succinyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N6-[(R)-dihydrolipoyl]-L-lysyl-[protein] + succinyl-CoA = N6-[(R)- S(8)-succinyldihydrolipoyl]-L-lysyl-[protein] + CoA
|
 |
 |
 |
 |
 |
N(6)-[(R)-dihydrolipoyl]-L-lysyl-[protein]
|
+
|
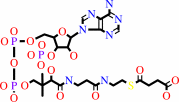 succinyl-CoA
succinyl-CoA
|
=
|
N(6)-[(R)- S(8)-succinyldihydrolipoyl]-L-lysyl-[protein]
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
E.C.4.1.1.71
- 2-oxoglutarate decarboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2-oxoglutarate + H+ = succinate semialdehyde + CO2
|
 |
 |
 |
 |
 |
 2-oxoglutarate
2-oxoglutarate
|
+
|
H(+)
|
=
|
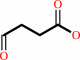 succinate semialdehyde
succinate semialdehyde
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Thiamine diphosphate
|
 |
 |
 |
 |
 |
Thiamine diphosphate
Bound ligand (Het Group name =
TD7)
matches with 78.79% similarity
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Chem Biol
18:1011-1020
(2011)
|
|
PubMed id:
|
|
|
|

|
| |
|
Functional plasticity and allosteric regulation of α-ketoglutarate decarboxylase in central mycobacterial metabolism.
|
|
T.Wagner,
M.Bellinzoni,
A.Wehenkel,
H.M.O'Hare,
P.M.Alzari.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

