 |
PDBsum entry 2iu8

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.191
- UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a UDP-3-O-[(3R)-3-hydroxyacyl]-alpha-D-glucosamine + a (3R)-hydroxyacyl- [ACP] = a UDP-2-N,3-O-bis[(3R)-3-hydroxyacyl]-alpha-D-glucosamine + holo- [ACP] + H+
|
 |
 |
 |
 |
 |
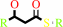 (3R)-3-hydroxyacyl-[acyl-carrier-protein]
(3R)-3-hydroxyacyl-[acyl-carrier-protein]
|
+
|
UDP-3-O-((3R)- hydroxyacyl)-alpha-D-glucosamine
|
=
|
UDP-2-N,3-O-bis((3R)-3-hydroxyacyl)- alpha-D-glucosamine
|
+
|
 holo-[acyl-carrier-protein]
holo-[acyl-carrier-protein]
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
104:4321-4326
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure and reactivity of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
L.Buetow,
T.K.Smith,
A.Dawson,
S.Fyffe,
W.N.Hunter.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The external layer of the Gram-negative bacterial outer membrane is primarily
composed of a protective, selectively permeable LPS. The biosynthesis of LPS
relies on UDP-3-O-acyl-glucosamine N-acyltransferase (LpxD), which transfers
3-hydroxy-arachidic acid from acyl carrier protein to the 2' amine of
UDP-3-O-myristoyl glucosamine in Chlamydia trachomatis. Our crystallographic
study reveals that LpxD is a homotrimer, each subunit of which is constructed
from a novel combination of an N-terminal uridine-binding domain, a core
lipid-binding domain, and a C-terminal helical extension. Highly conserved
residues dominate nucleotide binding. Phe-43 and Tyr-49 form pi-stacking
interactions with uracil, and Asn-46 and His-284 form hydrogen bonds with the
phosphate groups. These interactions place the glucosamine moiety at the
catalytic center formed by two adjacent subunits. Here His-247 and His-284
contribute to a mechanism involving nucleophilic attack by the amine of one
substrate on the carbonyl carbon of an acyl carrier protein thioester conjugate.
Serendipitously, our study reveals a fatty acid (FA) binding groove near the
catalytic center. MS elucidated the presence of a FA mixture binding to LpxD,
with palmitic acid the most prevalent. The placement of UDP-N-acetylglucosamine
and the FA provides details of N-acyltransferase ligand interactions and allows
for a description of structure and reactivity at an early stage of LPS assembly.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. Structure of LpxD. (A) Ribbon diagram of a subunit.
The UBD is yellow, the LBD is blue, loops are magenta, and the
HE is red. Selected elements of secondary structure are labeled,
and the coils are numbered. (B) The trimer. The view is parallel
to the noncrystallographic symmetry threefold axis. Subunits are
colored gray, wheat, and slate, and the domains of the gray
subunit are labeled. UDP-GlcNAc (complex II) is represented by
spheres, and palmitic acid is represented by sticks. The atoms
of the ligands are colored as follows: C, green; N, blue; O,
red; P, yellow. (C) Orthogonal view of the trimer. (D) Primary
and secondary structure.  -strands are depicted by
arrows, and -strands are depicted by
arrows, and  -helices are depicted
by cylinders. Colors are as described in A. Disordered residues
at the C terminus are marked by dots. Highly conserved residues
(>60% identity in 85 sequences) are highlighted in gray, and
strictly conserved residues (100% identity) are in black. Green
stars and circles indicate residues that interact with
UDP-GlcNAc and palmitic acid, respectively. Salmon boxes
represent sites of conditionally lethal point mutations in E.
coli and S. typhimurium LpxD. -helices are depicted
by cylinders. Colors are as described in A. Disordered residues
at the C terminus are marked by dots. Highly conserved residues
(>60% identity in 85 sequences) are highlighted in gray, and
strictly conserved residues (100% identity) are in black. Green
stars and circles indicate residues that interact with
UDP-GlcNAc and palmitic acid, respectively. Salmon boxes
represent sites of conditionally lethal point mutations in E.
coli and S. typhimurium LpxD.
|
 |
Figure 4.
Fig. 4. LpxD–FA complex. (A) Identification of bound FA.
GC-MS analysis, chain length, and saturation states are
indicated, and the key shows the relative percentages. (B)
Surface view of conserved residues with ligands depicted as
sticks. Palmitic acid is colored cyan, and UDP-GlcNAc is colored
according to atom type: C, white; N, blue; O, red. Conserved
residues are colored by type; basic residues are blue with the
exception of His-247 and His-284, which are colored green.
Acidic residues are red, aromatic residues are salmon, glycine
residues are yellow, polar residues (Asn, Gln, Ser, Thr, and
Cys) are slate blue, and aliphatic residues (Ala, Ile, Val, Leu,
Met, and Pro) are magenta. Colored residues that form part of
the FA and UDP-GlcNAc binding pockets include Gly-262, Gly-280,
Gly-265, Ala-246, Ile-263, Ala-264, Asp-240, and Gln-244.
|
 |
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Patarakul,
M.Lo,
and
B.Adler
(2010).
Global transcriptomic response of Leptospira interrogans serovar Copenhageni upon exposure to serum.
|
| |
BMC Microbiol,
10,
31.
|
 |
|
|
|
|
 |
X.Wang,
and
P.J.Quinn
(2010).
Lipopolysaccharide: Biosynthetic pathway and structure modification.
|
| |
Prog Lipid Res,
49,
97.
|
 |
|
|
|
|
 |
C.M.Bartling,
and
C.R.Raetz
(2009).
Crystal structure and acyl chain selectivity of Escherichia coli LpxD, the N-acyltransferase of lipid A biosynthesis.
|
| |
Biochemistry,
48,
8672-8683.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.R.Raetz,
Z.Guan,
B.O.Ingram,
D.A.Six,
F.Song,
X.Wang,
and
J.Zhao
(2009).
Discovery of new biosynthetic pathways: the lipid A story.
|
| |
J Lipid Res,
50,
S103-S108.
|
 |
|
|
|
|
 |
M.Demendi,
and
C.Creuzenet
(2009).
Cj1123c (PglD), a multifaceted acetyltransferase from Campylobacter jejuni.
|
| |
Biochem Cell Biol,
87,
469-483.
|
 |
|
|
|
|
 |
N.L.Ramsden,
L.Buetow,
A.Dawson,
L.A.Kemp,
V.Ulaganathan,
R.Brenk,
G.Klebe,
and
W.N.Hunter
(2009).
A structure-based approach to ligand discovery for 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase: a target for antimicrobial therapy.
|
| |
J Med Chem,
52,
2531-2542.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.W.Bainbridge,
L.Karimi-Naser,
R.Reife,
F.Blethen,
R.K.Ernst,
and
R.P.Darveau
(2008).
Acyl chain specificity of the acyltransferases LpxA and LpxD and substrate availability contribute to lipid A fatty acid heterogeneity in Porphyromonas gingivalis.
|
| |
J Bacteriol,
190,
4549-4558.
|
 |
|
|
|
|
 |
C.M.Bartling,
and
C.R.Raetz
(2008).
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
| |
Biochemistry,
47,
5290-5302.
|
 |
|
|
|
|
 |
N.B.Olivier,
and
B.Imperiali
(2008).
Crystal structure and catalytic mechanism of PglD from Campylobacter jejuni.
|
| |
J Biol Chem,
283,
27937-27946.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.H.Williams,
and
C.R.Raetz
(2007).
Structural basis for the acyl chain selectivity and mechanism of UDP-N-acetylglucosamine acyltransferase.
|
| |
Proc Natl Acad Sci U S A,
104,
13543-13550.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.R.Raetz,
C.M.Reynolds,
M.S.Trent,
and
R.E.Bishop
(2007).
Lipid A modification systems in gram-negative bacteria.
|
| |
Annu Rev Biochem,
76,
295-329.
|
 |
|
|
|
|
 |
D.M.Byers,
and
H.Gong
(2007).
Acyl carrier protein: structure-function relationships in a conserved multifunctional protein family.
|
| |
Biochem Cell Biol,
85,
649-662.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

