
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Wild type ssat- 1.7a structure
|
|
Structure:
|
 |
Diamine acetyltransferase 1. Chain: a. Synonym: spermidine/spermine n1, - acetyltransferase 1, ssat, ssat-1, putrescine acetyltransferase, polyamine n-acetyltransferase 1. Engineered: yes. Diamine acetyltransferase 1. Chain: b. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: sat. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.211
|
R-free:
|
0.241
|
|
|
Authors:
|
 |
M.C.Bewley,V.Graziano,J.S.Jiang,E.Matz,F.W.Studier,A.P.Pegg, C.S.Coleman,J.M.Flanagan,S.K.Burley,New York Sgx Research Center For Structural Genomics (Nysgxrc)
|
Key ref:

|
 |
M.C.Bewley
et al.
(2006).
Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target.
Proc Natl Acad Sci U S A,
103,
2063-2068.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
28-Sep-05
|
Release date:
|
17-Jan-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.2.3.1.57
- diamine N-acetyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an alkane-alpha,omega-diamine + acetyl-CoA = an N-acetylalkane- alpha,omega-diamine + CoA + H+
|
 |
 |
 |
 |
 |
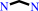 alkane-alpha,omega-diamine
alkane-alpha,omega-diamine
|
+
|
 acetyl-CoA
acetyl-CoA
|
=
|
N-acetylalkane- alpha,omega-diamine
|
+
|
 CoA
CoA
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
103:2063-2068
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target.
|
|
M.C.Bewley,
V.Graziano,
J.Jiang,
E.Matz,
F.W.Studier,
A.E.Pegg,
C.S.Coleman,
J.M.Flanagan.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Spermidine/spermine N(1)-acetyltransferase (SSAT) is a key enzyme in the control
of polyamine levels in human cells, as acetylation of spermidine and spermine
triggers export or degradation. Increased intracellular polyamine levels
accompany several types of cancers as well as other human diseases, and
compounds that affect the expression, activity, or stability of SSAT are being
explored as potential therapeutic drugs. We have expressed human SSAT from the
cloned cDNA in Escherichia coli and have determined high-resolution structures
of wild-type and mutant SSAT, as the free dimer and in binary and ternary
complexes with CoA, acetyl-CoA (AcCoA), spermine, and the inhibitor
N(1),N(11)-bis-(ethyl)-norspermine (BE-3-3-3). These structures show details of
binding sites for cofactor, substrates, and inhibitor and provide a framework to
understand enzymatic activity, mutations, and the action of potential drugs. Two
dimer conformations were observed: a symmetric form with two open surface
channels capable of binding substrate or cofactor, and an asymmetric form in
which only one of the surface channels appears capable of binding and
acetylating polyamines. SSAT was found to self-acetylate lysine-26 in the
presence of AcCoA and absence of substrate, a reaction apparently catalzyed by
AcCoA bound in the second channel of the asymmetric dimer. These unexpected and
intriguing complexities seem likely to have some as yet undefined role in
regulating SSAT activity or stability as a part of polyamine homeostasis.
Sequence signatures group SSAT with proteins that appear to have thialysine
N(epsilon)-acetyltransferase activity.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Space-filling view of the two channels on opposite
surfaces of the asymmetric dimer. The channels are at the dimer
interface, with chain A mostly to the upper left in the view of
channel 1 and to the lower right in the view of channel 2. The
two views are related by a 180° rotation about the
noncrystallographic 2-fold axis. Negatively charged residues
that line the channels are colored red, positively charged
residues are colored blue, and hydrophobic residues are colored
yellow. The suffix a or b on the amino acid designations
indicates whether the residue is from chain A or B. Arrows point
to the parts of channel 1 that house AcCoA and polyamine.
Channel 2 is occluded by residues 27-29 of chain B, which
prevents polyamine but not CoA from binding.
|
 |
Figure 4.
Fig. 4. Simulated-annealing omit maps showing the electron
density and interactions of bound CoA and BE-3-3-3. The electron
density corresponding to CoA (A and B) and BE-3-3-3 (C) is drawn
as light blue chicken wire. Oxygen atoms are red spheres, and
nitrogen atoms are blue spheres. (A) Interactions of CoA that
occur in both channel 1 and 2 of the asymmetric dimer. Backbone
nitrogens from P-loop residues G102, F103, and G104 interact
with the pyrophosphate moiety of CoA. R101, Y140, R142, and R143
side chains from the same monomer are drawn in ball-and-stick
representation. (B) Hydrogen bond network with chain A residues
that houses the pantetheine moiety of CoA in channel 1 of the
asymmetric dimer. Side-chain residues of 94 and 95 beyond C[
 ]have been omitted for
clarity. Hydrogen bonds are drawn as dashed lines. (C)
Interactions of BE-3-3-3 in channel 1 of the asymmetric dimer.
Ball-and-stick representations of residues from chain A and B of
the asymmetric dimer are drawn in purple and green, respectively. ]have been omitted for
clarity. Hydrogen bonds are drawn as dashed lines. (C)
Interactions of BE-3-3-3 in channel 1 of the asymmetric dimer.
Ball-and-stick representations of residues from chain A and B of
the asymmetric dimer are drawn in purple and green, respectively.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.B.Lee,
J.H.Park,
J.E.Folk,
J.A.Deck,
A.E.Pegg,
M.Sokabe,
C.S.Fraser,
and
M.H.Park
(2010).
Inactivation of eukaryotic initiation factor 5A (eIF5A) by specific acetylation of its hypusine residue by spermidine/spermine acetyltransferase 1 (SSAT1).
|
| |
Biochem J,
433,
205-213.
|
 |
|
|
|
|
 |
D.E.McCloskey,
S.Bale,
J.A.Secrist,
A.Tiwari,
T.H.Moss,
J.Valiyaveettil,
W.H.Brooks,
W.C.Guida,
A.E.Pegg,
and
S.E.Ealick
(2009).
New insights into the design of inhibitors of human S-adenosylmethionine decarboxylase: studies of adenine C8 substitution in structural analogues of S-adenosylmethionine.
|
| |
J Med Chem,
52,
1388-1407.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Zhou,
and
Z.Shang
(2009).
2D molecular graphics: a flattened world of chemistry and biology.
|
| |
Brief Bioinform,
10,
247-258.
|
 |
|
|
|
|
 |
R.A.Casero,
and
A.E.Pegg
(2009).
Polyamine catabolism and disease.
|
| |
Biochem J,
421,
323-338.
|
 |
|
|
|
|
 |
R.A.Casero,
and
L.J.Marton
(2007).
Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases.
|
| |
Nat Rev Drug Discov,
6,
373-390.
|
 |
|
|
|
|
 |
S.S.Hegde,
J.Chandler,
M.W.Vetting,
M.Yu,
and
J.S.Blanchard
(2007).
Mechanistic and structural analysis of human spermidine/spermine N1-acetyltransferase.
|
| |
Biochemistry,
46,
7187-7195.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.W.Han,
C.A.Bingman,
G.E.Wesenberg,
and
G.N.Phillips
(2006).
Crystal structure of Homo sapiens thialysine Nepsilon-acetyltransferase (HsSSAT2) in complex with acetyl coenzyme A.
|
| |
Proteins,
64,
288-293.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

