 |
PDBsum entry 1vpx

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of transaldolase (ec 2.2.1.2) (tm0295) from thermotoga maritima at 2.40 a resolution
|
|
Structure:
|
 |
Protein (transaldolase (ec 2.2.1.2)). Chain: a, b, c, d, e, f, g, h, i, j, k, l, m, n, o, p, q, r, s, t. Engineered: yes
|
|
Source:
|
 |
Thermotoga maritima. Organism_taxid: 2336. Gene: tm0295. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.199
|
R-free:
|
0.247
|
|
|
Authors:
|
 |
Joint Center For Structural Genomics (Jcsg)
|
|
Key ref:
|
 |
Joint center for structural genomics (jcsg)
Crystal structure of transaldolase (EC 2.2.1.2) (tm0295) from thermotoga maritima at 2.40 a resolution.
To be published,
.
|
 |
|
Date:
|
 |
|
23-Nov-04
|
Release date:
|
07-Dec-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9WYD1
(TAL_THEMA) -
Transaldolase from Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
218 a.a.
207 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.2.1.2
- transaldolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate = D-erythrose 4-phosphate + beta-D-fructose 6-phosphate
|
 |
 |
 |
 |
 |
D-sedoheptulose 7-phosphate
Bound ligand (Het Group name = )
matches with 60.00% similarity
|
+
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
=
|
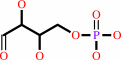 D-erythrose 4-phosphate
D-erythrose 4-phosphate
|
+
|
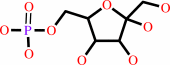 beta-D-fructose 6-phosphate
beta-D-fructose 6-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

