 |
PDBsum entry 1vkb

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.3.2.8
- gamma-glutamylamine cyclotransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
epsilon-(gamma-L-glutamyl)-L-lysine = 5-oxo-L-proline + L-lysine
|
 |
 |
 |
 |
 |
epsilon-(gamma-L-glutamyl)-L-lysine
|
=
|
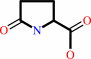 5-oxo-L-proline
5-oxo-L-proline
|
+
|
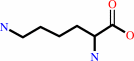 L-lysine
L-lysine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
61:1132-1136
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of a conserved hypothetical protein (gi: 13879369) from Mouse at 1.90 A resolution reveals a new fold.
|
|
H.E.Klock,
R.Schwarzenbacher,
Q.Xu,
D.McMullan,
P.Abdubek,
E.Ambing,
H.Axelrod,
T.Biorac,
J.M.Canaves,
H.J.Chiu,
A.M.Deacon,
M.DiDonato,
M.A.Elsliger,
A.Godzik,
C.Grittini,
S.K.Grzechnik,
J.Hale,
E.Hampton,
G.W.Han,
J.Haugen,
M.Hornsby,
L.Jaroszewski,
E.Koesema,
A.Kreusch,
P.Kuhn,
M.D.Miller,
K.Moy,
E.Nigoghossian,
J.Paulsen,
K.Quijano,
R.Reyes,
C.Rife,
E.Sims,
G.Spraggon,
R.C.Stevens,
H.van den Bedem,
J.Velasquez,
J.Vincent,
A.White,
G.Wolf,
K.O.Hodgson,
J.Wooley,
S.A.Lesley,
I.A.Wilson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Crystal structure of 13879369 from Mouse. (A) Ribbon
diagram color-coded from N-terminus (blue) to C-terminus (red)
showing the domain organization. Helices H1-H4 and  -strands -strands
 1- 1-
 7
are indicated. The partial disulfide bond between Cys35 and
Cys62 is shown in ball-and-stick. (B) Diagram showing the
secondary structure elements in 13879369 superimposed on its
primary sequence. The 7
are indicated. The partial disulfide bond between Cys35 and
Cys62 is shown in ball-and-stick. (B) Diagram showing the
secondary structure elements in 13879369 superimposed on its
primary sequence. The  -helices, -helices,
 -strands, -strands,
 -bulges, -bulges,
 -turns
and disordered regions (dots) are indicated. The -turns
and disordered regions (dots) are indicated. The  -sheet
strands are indicated by a red A and B. -sheet
strands are indicated by a red A and B.  -Hairpins
are depicted as red loops. -Hairpins
are depicted as red loops.
|
 |
Figure 2.
Figure 2. (A) Protein 13879369 in surface representation
showing the putative active site cavity with neighboring
residues in sticks. Two formic acid and three water molecules
located inside the cavity are depicted in sticks and red balls,
respectively. (B) Superposition of 13879369 (blue and red) and
Ytfp (gray) in ribbon representation. Regions where backbones
deviate by RMSD > 4 Å are colored red in 13879369. The two
formates shown in red and yellow spheres indicate the position
of the putative active site cavity.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2005,
61,
1132-1136)
copyright 2005.
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
P.Serrano,
B.Pedrini,
M.Geralt,
K.Jaudzems,
B.Mohanty,
R.Horst,
T.Herrmann,
M.A.Elsliger,
I.A.Wilson,
and
K.Wüthrich
(2010).
Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
66,
1393-1405.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Bae,
C.A.Bingman,
D.J.Aceti,
and
G.N.Phillips
(2008).
Crystal structure of Homo sapiens protein LOC79017.
|
| |
Proteins,
70,
588-591.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.M.Aramini,
Y.J.Huang,
G.V.Swapna,
J.R.Cort,
P.K.Rajan,
R.Xiao,
R.Shastry,
T.B.Acton,
J.Liu,
B.Rost,
M.A.Kennedy,
and
G.T.Montelione
(2007).
Solution NMR structure of Escherichia coli ytfP expands the structural coverage of the UPF0131 protein domain family.
|
| |
Proteins,
68,
789-795.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.M.Llewellyn,
Y.Li,
and
J.B.Spencer
(2007).
Biosynthesis of butirosin: transfer and deprotection of the unique amino acid side chain.
|
| |
Chem Biol,
14,
379-386.
|
 |
|
|
|
|
 |
B.L.Lytle,
F.C.Peterson,
E.M.Tyler,
C.L.Newman,
D.A.Vinarov,
J.L.Markley,
and
B.F.Volkman
(2006).
Solution structure of Arabidopsis thaliana protein At5g39720.1, a member of the AIG2-like protein family.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
490-493.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

