 |
PDBsum entry 1v1j

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of type ii dehydroquintae dehydratase from streptomyces coelicolor in complex with 3-fluoro
|
|
Structure:
|
 |
3-dehydroquinate dehydratase. Chain: a, b, c, d, e, f, g, h, i, j, k, l. Synonym: 3-dehydroquinase, type ii dhqase. Engineered: yes
|
|
Source:
|
 |
Streptomyces coelicolor. Organism_taxid: 1902. Expressed in: escherichia coli. Expression_system_taxid: 511693.
|
|
Biol. unit:
|
 |
Dodecamer (from PDB file)
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.217
|
R-free:
|
0.238
|
|
|
Authors:
|
 |
A.W.Roszak,J.R.Coggins,A.J.Lapthorn
|
|
Key ref:
|
 |
M.Frederickson
et al.
(2004).
(1R,4S,5R)-3-Fluoro-1,4,5-trihydroxy-2-cyclohexene-1-carboxylic acid: the fluoro analogue of the enolate intermediate in the reaction catalyzed by type II dehydroquinases.
Org Biomol Chem,
2,
1592-1596.
PubMed id:
|
 |
|
Date:
|
 |
|
16-Apr-04
|
Release date:
|
26-Jan-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P15474
(AROQ_STRCO) -
3-dehydroquinate dehydratase from Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
157 a.a.
150 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.10
- 3-dehydroquinate dehydratase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Shikimate and Chorismate Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3-dehydroquinate = 3-dehydroshikimate + H2O
|
 |
 |
 |
 |
 |
3-dehydroquinate
Bound ligand (Het Group name = )
matches with 85.71% similarity
|
=
|
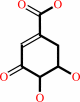 3-dehydroshikimate
3-dehydroshikimate
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Org Biomol Chem
2:1592-1596
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
(1R,4S,5R)-3-Fluoro-1,4,5-trihydroxy-2-cyclohexene-1-carboxylic acid: the fluoro analogue of the enolate intermediate in the reaction catalyzed by type II dehydroquinases.
|
|
M.Frederickson,
A.W.Roszak,
J.R.Coggins,
A.J.Lapthorn,
C.Abell.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The fluoro analogue of the enolate intermediate in the reaction catalyzed by
type II dehydroquinases has been prepared from naturally occurring (-)-quinic
acid over seven steps and has been shown to be the most potent inhibitor
reported to date of the type II enzyme from Mycobacterium tuberculosis.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Paz,
L.Tizón,
J.M.Otero,
A.L.Llamas-Saiz,
G.C.Fox,
M.J.van Raaij,
H.Lamb,
A.R.Hawkins,
A.J.Lapthorn,
L.Castedo,
and
C.González-Bello
(2011).
Tetrahydrobenzothiophene Derivatives: Conformationally Restricted Inhibitors of Type II Dehydroquinase.
|
| |
ChemMedChem,
6,
266-272.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Kumar,
M.I.Siddiqi,
and
S.Miertus
(2010).
New molecular scaffolds for the design of Mycobacterium tuberculosis type II dehydroquinase inhibitors identified using ligand and receptor based virtual screening.
|
| |
J Mol Model,
16,
693-712.
|
 |
|
|
|
|
 |
A.Peón,
J.M.Otero,
L.Tizón,
V.F.Prazeres,
A.L.Llamas-Saiz,
G.C.Fox,
M.J.van Raaij,
H.Lamb,
A.R.Hawkins,
F.Gago,
L.Castedo,
and
C.González-Bello
(2010).
Understanding the key factors that control the inhibition of type II dehydroquinase by (2R)-2-benzyl-3-dehydroquinic acids.
|
| |
ChemMedChem,
5,
1726-1733.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Trapani,
G.Schoehn,
J.Navaza,
and
C.Abergel
(2010).
Macromolecular crystal data phased by negative-stained electron-microscopy reconstructions.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
514-521.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.González-Bello,
and
L.Castedo
(2007).
Progress in type II dehydroquinase inhibitors: from concept to practice.
|
| |
Med Res Rev,
27,
177-208.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

