 |
PDBsum entry 1ro5

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Signaling protein
|
PDB id
|
|

|

|
1ro5
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.184
- acyl-homoserine-lactone synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a fatty acyl-[ACP] + S-adenosyl-L-methionine = an N-acyl-L-homoserine lactone + S-methyl-5'-thioadenosine + holo-[ACP] + H+
|
 |
 |
 |
 |
 |
fatty acyl-[ACP]
|
+
|
 S-adenosyl-L-methionine
S-adenosyl-L-methionine
|
=
|
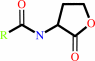 N-acyl-L-homoserine lactone
N-acyl-L-homoserine lactone
|
+
|
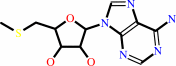 S-methyl-5'-thioadenosine
S-methyl-5'-thioadenosine
|
+
|
holo-[ACP]
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Mol Microbiol
53:1135-1146
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of the Pseudomonas aeruginosa acyl-homoserinelactone synthase LasI.
|
|
T.A.Gould,
H.P.Schweizer,
M.E.Churchill.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The LasI/LasR quorum-sensing system plays a pivotal role in virulence gene
regulation of the opportunistic human pathogen, Pseudomonas aeruginosa. Here we
report the crystal structure of the acyl-homoserine lactone (AHL) synthase LasI
that produces 3-oxo-C12-AHL from the substrates 3-oxo-C12-acyl-carrier protein
(acyl-ACP) and S-adenosyl-L-methionine. The LasI six-stranded beta sheet
platform, buttressed by three alpha helices, forms a V-shaped substrate-binding
cleft that leads to a tunnel passing through the enzyme that can accommodate the
acyl-chain of acyl-ACP. This tunnel places no apparent restriction on acyl-chain
length, in contrast to a restrictive hydrophobic pocket seen in the AHL-synthase
EsaI. Interactions of essential conserved N-terminal residues, Arg23, Phe27 and
Trp33, suggest that the N-terminus forms an enclosed substrate-binding pocket
for S-adenosyl-L-methionine. Analysis of AHL-synthase surface residues
identified a binding site for acyl-ACP, a role that was supported by in vivo
reporter assay analysis of the mutated residues, including Arg154 and Lys150.
This structure and the novel explanation of AHL-synthase acyl-chain-length
selectivity promise to guide the design of Pseudomonas aeruginosa-specific
quorum-sensing inhibitors as antibacterial agents.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.C.Kwan,
T.Meickle,
D.Ladwa,
M.Teplitski,
V.Paul,
and
H.Luesch
(2011).
Lyngbyoic acid, a "tagged" fatty acid from a marine cyanobacterium, disrupts quorum sensing in Pseudomonas aeruginosa.
|
| |
Mol Biosyst,
7,
1205-1216.
|
 |
|
|
|
|
 |
M.A.Savka,
P.T.Le,
and
T.J.Burr
(2011).
LasR receptor for detection of long-chain quorum-sensing signals: identification of N-acyl-homoserine lactones encoded by the avsI locus of Agrobacterium vitis.
|
| |
Curr Microbiol,
62,
101-110.
|
 |
|
|
|
|
 |
J.S.Dickschat
(2010).
Quorum sensing and bacterial biofilms.
|
| |
Nat Prod Rep,
27,
343-369.
|
 |
|
|
|
|
 |
M.Tizzano,
B.D.Gulbransen,
A.Vandenbeuch,
T.R.Clapp,
J.P.Herman,
H.M.Sibhatu,
M.E.Churchill,
W.L.Silver,
S.C.Kinnamon,
and
T.E.Finger
(2010).
Nasal chemosensory cells use bitter taste signaling to detect irritants and bacterial signals.
|
| |
Proc Natl Acad Sci U S A,
107,
3210-3215.
|
 |
|
|
|
|
 |
N.Bhargava,
P.Sharma,
and
N.Capalash
(2010).
Quorum sensing in Acinetobacter: an emerging pathogen.
|
| |
Crit Rev Microbiol,
36,
349-360.
|
 |
|
|
|
|
 |
P.K.Kambam,
D.T.Eriksen,
J.Lajoie,
D.J.Sayut,
and
L.Sun
(2009).
Altering the substrate specificity of RhlI by directed evolution.
|
| |
Chembiochem,
10,
553-558.
|
 |
|
|
|
|
 |
W.L.Ng,
and
B.L.Bassler
(2009).
Bacterial quorum-sensing network architectures.
|
| |
Annu Rev Genet,
43,
197-222.
|
 |
|
|
|
|
 |
A.Takaya,
F.Tabuchi,
H.Tsuchiya,
E.Isogai,
and
T.Yamamoto
(2008).
Negative regulation of quorum-sensing systems in Pseudomonas aeruginosa by ATP-dependent Lon protease.
|
| |
J Bacteriol,
190,
4181-4188.
|
 |
|
|
|
|
 |
G.D.Geske,
J.C.O'Neill,
and
H.E.Blackwell
(2008).
Expanding dialogues: from natural autoinducers to non-natural analogues that modulate quorum sensing in Gram-negative bacteria.
|
| |
Chem Soc Rev,
37,
1432-1447.
|
 |
|
|
|
|
 |
J.S.Cisar,
and
D.S.Tan
(2008).
Small molecule inhibition of microbial natural product biosynthesis-an emerging antibiotic strategy.
|
| |
Chem Soc Rev,
37,
1320-1329.
|
 |
|
|
|
|
 |
M.Cooley,
S.R.Chhabra,
and
P.Williams
(2008).
N-Acylhomoserine lactone-mediated quorum sensing: a twist in the tail and a blow for host immunity.
|
| |
Chem Biol,
15,
1141-1147.
|
 |
|
|
|
|
 |
P.K.Kambam,
D.J.Sayut,
Y.Niu,
D.T.Eriksen,
and
L.Sun
(2008).
Directed evolution of LuxI for enhanced OHHL production.
|
| |
Biotechnol Bioeng,
101,
263-272.
|
 |
|
|
|
|
 |
C.E.White,
and
S.C.Winans
(2007).
Cell-cell communication in the plant pathogen Agrobacterium tumefaciens.
|
| |
Philos Trans R Soc Lond B Biol Sci,
362,
1135-1148.
|
 |
|
|
|
|
 |
P.Williams,
K.Winzer,
W.C.Chan,
and
M.Cámara
(2007).
Look who's talking: communication and quorum sensing in the bacterial world.
|
| |
Philos Trans R Soc Lond B Biol Sci,
362,
1119-1134.
|
 |
|
|
|
|
 |
R.Van Houdt,
M.Givskov,
and
C.W.Michiels
(2007).
Quorum sensing in Serratia.
|
| |
FEMS Microbiol Rev,
31,
407-424.
|
 |
|
|
|
|
 |
W.Nasser,
and
S.Reverchon
(2007).
New insights into the regulatory mechanisms of the LuxR family of quorum sensing regulators.
|
| |
Anal Bioanal Chem,
387,
381-390.
|
 |
|
|
|
|
 |
G.Hao,
and
T.J.Burr
(2006).
Regulation of long-chain N-acyl-homoserine lactones in Agrobacterium vitis.
|
| |
J Bacteriol,
188,
2173-2183.
|
 |
|
|
|
|
 |
M.E.Churchill
(2006).
A new GNAT in bacterial signaling?
|
| |
Structure,
14,
1342-1344.
|
 |
|
|
|
|
 |
R.A.Scott,
J.Weil,
P.T.Le,
P.Williams,
R.G.Fray,
S.B.von Bodman,
and
M.A.Savka
(2006).
Long- and short-chain plant-produced bacterial N-acyl-homoserine lactones become components of phyllosphere, rhizosphere, and soil.
|
| |
Mol Plant Microbe Interact,
19,
227-239.
|
 |
|
|
|
|
 |
R.M.Van Wagoner,
and
J.Clardy
(2006).
FeeM, an N-acyl amino acid synthase from an uncultured soil microbe: structure, mechanism, and acyl carrier protein binding.
|
| |
Structure,
14,
1425-1435.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.A.Gould,
J.Herman,
J.Krank,
R.C.Murphy,
and
M.E.Churchill
(2006).
Specificity of acyl-homoserine lactone synthases examined by mass spectrometry.
|
| |
J Bacteriol,
188,
773-783.
|
 |
|
|
|
|
 |
C.Farah,
M.Vera,
D.Morin,
D.Haras,
C.A.Jerez,
and
N.Guiliani
(2005).
Evidence for a functional quorum-sensing type AI-1 system in the extremophilic bacterium Acidithiobacillus ferrooxidans.
|
| |
Appl Environ Microbiol,
71,
7033-7040.
|
 |
|
|
|
|
 |
C.M.Waters,
and
B.L.Bassler
(2005).
Quorum sensing: cell-to-cell communication in bacteria.
|
| |
Annu Rev Cell Dev Biol,
21,
319-346.
|
 |
|
|
|
|
 |
G.Brader,
S.Sjöblom,
H.Hyytiäinen,
K.Sims-Huopaniemi,
and
E.T.Palva
(2005).
Altering substrate chain length specificity of an acylhomoserine lactone synthase in bacterial communication.
|
| |
J Biol Chem,
280,
10403-10409.
|
 |
|
|
|
|
 |
K.L.Visick,
and
C.Fuqua
(2005).
Decoding microbial chatter: cell-cell communication in bacteria.
|
| |
J Bacteriol,
187,
5507-5519.
|
 |
|
|
|
|
 |
P.Z.Kozbial,
and
A.R.Mushegian
(2005).
Natural history of S-adenosylmethionine-binding proteins.
|
| |
BMC Struct Biol,
5,
19.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

