 |
PDBsum entry 1r56

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1r56
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Uncomplexed urate oxidase from aspergillus flavus
|
|
Structure:
|
 |
Uricase. Chain: a, b, c, d, e, f, g, h. Synonym: urate oxidase. Ec: 1.7.3.3
|
|
Source:
|
 |
Aspergillus flavus. Organism_taxid: 5059
|
|
Biol. unit:
|
 |
 Tetramer (from
)
Tetramer (from
)
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.162
|
R-free:
|
0.214
|
|
|
Authors:
|
 |
P.Retailleau,N.Colloc'H,T.Prange
|
Key ref:

|
 |
P.Retailleau
et al.
(2004).
Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: a reassignment of the active-site binding mode.
Acta Crystallogr D Biol Crystallogr,
60,
453-462.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
09-Oct-03
|
Release date:
|
02-Mar-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q00511
(URIC_ASPFL) -
Uricase from Aspergillus flavus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
302 a.a.
295 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.7.3.3
- factor independent urate hydroxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP Catabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
urate + O2 + H2O = 5-hydroxyisourate + H2O2
|
 |
 |
 |
 |
 |
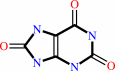 urate
urate
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
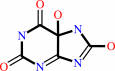 5-hydroxyisourate
5-hydroxyisourate
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Copper
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
60:453-462
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: a reassignment of the active-site binding mode.
|
|
P.Retailleau,
N.Colloc'h,
D.Vivarès,
F.Bonneté,
B.Castro,
M.El-Hajji,
J.P.Mornon,
G.Monard,
T.Prangé.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
High-resolution X-ray structures of the complexes of Aspergillus flavus urate
oxidase (Uox) with three inhibitors, 8-azaxanthin (AZA), 9-methyl uric acid
(MUA) and oxonic acid (OXC), were determined in an orthorhombic space group
(I222). In addition, the ligand-free enzyme was also crystallized in a
monoclinic form (P2(1)) and its structure determined. Higher accuracy in the
three new enzyme-inhibitor complex structures (Uox-AZA, Uox-MUA and Uox-OXC)
with respect to the previously determined structure of Uox-AZA (PDB code 1uox)
leads to a reversed position of the inhibitor in the active site of the enzyme.
The corrected anchoring of the substrate (uric acid) allows an improvement in
the understanding of the enzymatic mechanism of urate oxidase.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 Schematic degradation pathway of uric acid to allantoin
through intermediate (2) (5-hydroxyisourate). Only the first
step is catalyzed by the enzyme, with H[2]O[2] as the by-product.
|
 |
Figure 7.
Figure 7 Revised position of 8-azaxanthin relative to the active
site of Uox. (a) Original published location of the AZA
inhibitor in [212]1uox ; (b) the revised orientation of AZA in
the new refined structure (1.8 Å). Both orientations are
superimposed on the respective final 2|F[obs]| - |F[calc]|
electron-density maps contoured around the inhibitor at 1.75
[213][sigma] .
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2004,
60,
453-462)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.Gabison,
C.Chopard,
N.Colloc'h,
F.Peyrot,
B.Castro,
M.E.Hajji,
M.Altarsha,
G.Monard,
M.Chiadmi,
and
T.Prangé
(2011).
X-ray, ESR, and quantum mechanics studies unravel a spin well in the cofactor-less urate oxidase.
|
| |
Proteins,
79,
1964-1976.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Pompidor,
O.Maury,
J.Vicat,
and
R.Kahn
(2010).
A dipicolinate lanthanide complex for solving protein structures using anomalous diffraction.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
762-769.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Collings,
Y.Watier,
M.Giffard,
S.Dagogo,
R.Kahn,
F.Bonneté,
J.P.Wright,
A.N.Fitch,
and
I.Margiolaki
(2010).
Polymorphism of microcrystalline urate oxidase from Aspergillus flavus.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
539-548.
|
 |
|
|
|
|
 |
L.Gabison,
M.Chiadmi,
M.El Hajji,
B.Castro,
N.Colloc'h,
and
T.Prangé
(2010).
Near-atomic resolution structures of urate oxidase complexed with its substrate and analogues: the protonation state of the ligand.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
714-724.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.S.Till,
and
G.M.Ullmann
(2010).
McVol - a program for calculating protein volumes and identifying cavities by a Monte Carlo algorithm.
|
| |
J Mol Model,
16,
419-429.
|
 |
|
|
|
|
 |
B.Sankaran,
S.A.Bonnett,
K.Shah,
S.Gabriel,
R.Reddy,
P.Schimmel,
D.A.Rodionov,
V.de Crécy-Lagard,
J.D.Helmann,
D.Iwata-Reuyl,
and
M.A.Swairjo
(2009).
Zinc-independent folate biosynthesis: genetic, biochemical, and structural investigations reveal new metal dependence for GTP cyclohydrolase IB.
|
| |
J Bacteriol,
191,
6936-6949.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Oksanen,
M.P.Blakeley,
F.Bonneté,
M.T.Dauvergne,
F.Dauvergne,
and
M.Budayova-Spano
(2009).
Large crystal growth by thermal control allows combined X-ray and neutron crystallographic studies to elucidate the protonation states in Aspergillus flavus urate oxidase.
|
| |
J R Soc Interface,
6,
S599-S610.
|
 |
|
|
|
|
 |
Z.Liu,
D.Lu,
J.Li,
W.Chen,
and
Z.Liu
(2009).
Strengthening intersubunit hydrogen bonds for enhanced stability of recombinant urate oxidase from Aspergillus flavus: molecular simulations and experimental validation.
|
| |
Phys Chem Chem Phys,
11,
333-340.
|
 |
|
|
|
|
 |
J.E.Spoonamore,
S.A.Roberts,
A.Heroux,
and
V.Bandarian
(2008).
Structure of a 6-pyruvoyltetrahydropterin synthase homolog from Streptomyces coelicolor.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
875-879.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Gabison,
T.Prangé,
N.Colloc'h,
M.El Hajji,
B.Castro,
and
M.Chiadmi
(2008).
Structural analysis of urate oxidase in complex with its natural substrate inhibited by cyanide: mechanistic implications.
|
| |
BMC Struct Biol,
8,
32.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

