 |
PDBsum entry 1o5h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.3.1.4
- formimidoyltetrahydrofolate cyclodeaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Folate Coenzymes
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5-formimidoyltetrahydrofolate + 2 H+ = (6R)-5,10- methenyltetrahydrofolate + NH4+
|
 |
 |
 |
 |
 |
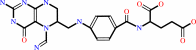 5-formimidoyltetrahydrofolate
5-formimidoyltetrahydrofolate
|
+
|
2
×
H(+)
|
=
|
(6R)-5,10- methenyltetrahydrofolate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
58:976-981
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of a formiminotetrahydrofolate cyclodeaminase (TM1560) from Thermotoga maritima at 2.80 A resolution reveals a new fold.
|
|
Q.Xu,
R.Schwarzenbacher,
D.McMullan,
P.Abdubek,
E.Ambing,
T.Biorac,
J.M.Canaves,
H.J.Chiu,
X.Dai,
A.M.Deacon,
M.DiDonato,
M.A.Elsliger,
A.Godzik,
C.Grittini,
S.K.Grzechnik,
E.Hampton,
M.Hornsby,
L.Jaroszewski,
H.E.Klock,
E.Koesema,
A.Kreusch,
P.Kuhn,
S.A.Lesley,
I.Levin,
M.D.Miller,
A.Morse,
K.Moy,
J.Ouyang,
R.Page,
K.Quijano,
R.Reyes,
A.Robb,
E.Sims,
G.Spraggon,
R.C.Stevens,
H.van den Bedem,
J.Velasquez,
J.Vincent,
F.von Delft,
X.Wang,
B.West,
A.White,
G.Wolf,
O.Zagnitko,
K.O.Hodgson,
J.Wooley,
I.A.Wilson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. (A) Scheme for the reaction carried out by FTCD_CD
(EC 4.3.1.4), which catalyzes the cyclization of the formimino
group of N^5-formimidoyltetrahydrofolate, yielding
N^5,N^10-methenyltetrahydrofolate and ammonia. (B) Crystal
structure of TM1560. Ribbon diagram of T. maritima FTCD_CD
color-coded from N-terminus (blue) to C-terminus (red) showing
the domain organization.  elices
H1-H6 are indicated. (C) The TM1560 dimer view is shown along
(above) and normal (below) to its two-fold axis. (D) Diagram
showing the secondary structure elements in TM1560 superimposed
on its primary sequence. The elices
H1-H6 are indicated. (C) The TM1560 dimer view is shown along
(above) and normal (below) to its two-fold axis. (D) Diagram
showing the secondary structure elements in TM1560 superimposed
on its primary sequence. The  -helices,
3[10]-helices, -helices,
3[10]-helices,  -bulges,
and -bulges,
and  -turns
are indicated. Disordered regions are depicted by a dashed line
with the corresponding sequence in brackets. -turns
are indicated. Disordered regions are depicted by a dashed line
with the corresponding sequence in brackets.
|
 |
Figure 2.
Figure 2. (A) Superposition of TM1560 (gray) and invertase
inhibitor Nt-Cif from Tobacco (PDB code: 1rj1, magenta). (B) The
putative active site pocket in surface representation. Residues
are colored according to sequence conservation, where green is
conserved and white is nonconserved. The manually docked ligand
N^5, N^10-methenyltetrahydrofolate (MTHF) is shown in stick
representation (yellow). (C) Same orientation as (B), but shown
in ribbon representation, with conserved residues in the
putative active site of TM1560 and the manually docked MTHF
ligand in stick representation. Monomers 1 and 2 in the dimer
are shown in blue and gray, respectively.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2005,
58,
976-981)
copyright 2005.
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

